Abstract
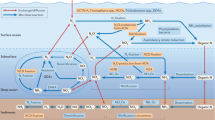
The role of macroalgae in Blue Carbon assessments has been controversial, partially due to uncertainties about the fate of exported macroalgae. Available evidence suggests that macroalgae are exported to reach the open ocean and the deep sea. Nevertheless, this evidence lacks systematic assessment. Here, we provide robust evidence of macroalgal export beyond coastal habitats. We used metagenomes and metabarcodes from the global expeditions Tara Oceans and Malaspina 2010 Circumnavigation. We discovered macroalgae worldwide at up to 5,000 km from coastal areas. We found 24 orders, most of which belong to the phylum Rhodophyta. The diversity of macroalgae was similar across oceanic regions, although the assemblage composition differed. The South Atlantic Ocean presented the highest macroalgal diversity, whereas the Red Sea was the least diverse region. The abundance of macroalgae sequences attenuated exponentially with depth at a rate of 37.3% km−1, and only 24% of macroalgae available at the surface were expected to reach the seafloor at a depth of 4,000 m. Our findings indicate that macroalgae are exported across the open and the deep ocean, suggesting that macroalgae may be an important source of allochthonous carbon, and their contribution should be considered in Blue Carbon assessments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study can be found in ref. 20 (Tara Oceans metagenomes; https://doi.org/10.1038/sdata.2015.23), and ref. 57 (Tara Oceans 18S rDNA V9 metabarcodes; https://doi.org/10.1126/science.1261605) and Zenodo (Malaspina metagenomes; https://doi.org/10.5281/zenodo.2596829)21.
References
Duarte, C. M. & Cebrián, J. The fate of marine autotrophic production. Limnol. Oceanogr. 41, 1758–1766 (1996).
Duarte, C. M. & Krause-Jensen, D. Export from seagrass meadows contributes to marine carbon sequestration. Front. Mar. Sci. 4, 13 (2017).
McLeod, E. et al. A blueprint for blue carbon: toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Front. Ecol. Environ. 9, 552–560 (2011).
Fourqurean, J. W. et al. Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5, 505–509 (2012).
Duarte, C. M., Kennedy, H., Marbà, N. & Hendriks, I. Assessing the capacity of seagrass meadows for carbon burial: current limitations and future strategies. Ocean Coast. Manage. 83, 32–38 (2013).
Donato, D. C. et al. Mangroves among the most carbon-rich forests in the tropics. Nat. Geosci. 4, 293–297 (2011).
Krause-Jensen, D. & Duarte, C. M. Substantial role of macroalgae in marine carbon sequestration. Nat. Geosci. 9, 737–742 (2016).
Krause-Jensen, D. et al. Sequestration of macroalgal carbon: the elephant in the Blue Carbon room. Biol. Lett. 14, 20180236 (2018).
Duarte, C. M. Reviews and syntheses: hidden forests, the role of vegetated coastal habitats in the ocean carbon budget. Biogeosciences 14, 301–310 (2017).
Garden, C. J. & Smith, A. M. Voyages of seaweeds: the role of macroalgae in sediment transport. Sediment. Geol. 318, 1–9 (2015).
Kloareg, B. & Quatrano, R. S. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. 26, 259–315 (1988).
Barrón, C., Apostolaki, E. T. & Duarte, C. M. Dissolved organic carbon fluxes by seagrass meadows and macroalgal beds. Front. Mar. Sci. 1, 42 (2014).
Krumhansl, K. A. & Scheibling, R. E. Production and fate of kelp detritus. Mar. Ecol. Prog. Ser. 467, 281–302 (2012).
Landenmark, H. K. E., Forgan, D. H. & Cockell, C. S. An estimate of the total DNA in the biosphere. PLoS Biol. 13, e1002168 (2015).
Karsenti, E. et al. A holistic approach to marine eco-systems biology. PLoS Biol. 9, e1001177 (2011).
Duarte, C. M. Seafaring in the 21st century: the Malaspina 2010 Circumnavigation Expedition. Limnol. Oceanogr. 24, 11–14 (2015).
Salazar, G. et al. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608 (2016).
Hingamp, P. et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 7, 1678–1695 (2013).
De Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
Pesant, S. et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2, 150023 (2015).
Sánchez, P. et al. Dataset: Common photosynthetic enzymes from 174 metagenomes from the Malaspina Expedition 2010. Supplement to: Ortega et al. (2019). Important contribution of macroalgae to oceanic carbon sequestration. Zenodo https://doi.org/10.5281/zenodo.2596829 (2019).
Guiry, M. D. How many species of algae are there? J. Phycol. 48, 1057–1063 (2012).
Cock, J. M. et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621 (2010).
Collins, R. A. et al. Persistence of environmental DNA in marine systems. Commun. Biol. 1, 185 (2018).
Thomsen, P. F. et al. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7, e41732 (2012).
Roux, S., Enault, F., le Bronner, G. & Debroas, D. Comparison of 16S rRNA and protein-coding genes as molecular markers for assessing microbial diversity (Bacteria and Archaea) in ecosystems. FEMS Microbiol. Ecol. 78, 617–628 (2011).
Seckbach, J. & Chapman, D. J. Red Algae in the Genomic Age (Springer, 2010).
Guiry, M. D. & Guiry, G. M. AlgaeBase (National Univ. Ireland, 2013); http://www.algaebase.org
Krause-Jensen, D. & Duarte, C. M. Expansion of vegetated coastal ecosystems in the future Arctic. Front. Mar. Sci. 1, 77 (2014).
Kaehler, S., Pakhomov, E. A., Kalin, R. M. & Davis, S. Trophic importance of kelp-derived suspended particulate matter in a through-flow sub-Antarctic system. Mar. Ecol. Prog. Ser. 316, 17–22 (2006).
Kelaher, B. P., Coleman, M. A. & Bishop, M. J. Ocean warming, but not acidification, accelerates seagrass decomposition under near-future climate scenarios. Mar. Ecol. Prog. Ser. 605, 103–110 (2018).
Cózar, A. et al. The Arctic Ocean as a dead end for floating plastics in the North Atlantic branch of the Thermohaline Circulation. Sci. Adv. 3, e1600582 (2017).
Martin, J. H., Knauer, G. A., Karl, D. M. & Broenkow, W. VERTEX: carbon cycling in the northeast Pacific. Deep Sea Res. Pt II 34, 267–285 (1987).
Enríquez, S., Duarte, C. M. & Sand-Jensen, K. A. J. Patterns in decomposition rates among photosynthetic organisms: the importance of detritus C:N:P content. Oecologia 94, 457–471 (1993).
Carpenter, E. J. & Cox, J. L. Production of pelagic Sargassum and a blue‐green epiphyte in the western Sargasso Sea. Limnol. Oceanogr. 19, 429–436 (1974).
Woodborne, M. W., Rogers, J. & Jarman, N. The geological significance of kelp-rafted rock along the west coast of South Africa. Geo-Mar. Lett. 9, 109–118 (1989).
Garden, C. J., Currie, K., Fraser, C. I. & Waters, J. M. Rafting dispersal constrained by an oceanographic boundary. Mar. Ecol. Prog. Ser. 501, 297–302 (2014).
Gattuso, J. P. et al. Light availability in the coastal ocean: impact on the distribution of benthic photosynthetic organisms and contribution to primary production. Biogeosciences 3, 895–959 (2006).
Littler, M. M., Littler, D. S., Blair, S. M. & Norris, J. N. Deepest known plant life discovered on an uncharted seamount. Science 227, 57–59 (1985).
Trevathan-Tackett, S. M. et al. Comparison of marine macrophytes for their contributions to blue carbon sequestration. Ecology 96, 3043–3057 (2015).
Percival, E. The polysaccharides of green, red and brown seaweeds: their basic structure, biosynthesis and function. Br. Phycol. 14, 103–117 (1979).
Shukla, P. S., Borza, T., Critchley, A. T. & Prithiviraj, B. Carrageenans from red seaweeds as promoters of growth and elicitors of defense response in plants. Front. Mar. Sci. 3, 81 (2016).
Berteau, O. & Mulloy, B. Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology 13, 29R–40R (2003).
Herzog, H., Caldeira, K. & Reilly, J. An issue of permanence: assessing the effectiveness of temporary carbon storage. Clim. Change 59, 293–310 (2003).
Robinson, C. et al. Mesopelagic zone ecology and biogeochemistry—a synthesis. Deep Sea Res. Pt II 57, 1504–1518 (2010).
Dierssen, H. M., Zimmerman, R. C., Drake, L. A. & Burdige, D. J. Potential export of unattached benthic macroalgae to the deep sea through wind-driven Langmuir circulation. Geophys. Res. Lett. 36, L04602 (2009).
De Leo, F. C., Smith, C. R., Rowden, A. A., Bowden, D. A. & Clark, M. R. Submarine canyons: hotspots of benthic biomass and productivity in the deep sea. Proc. R. Soc. Lond. B 277, 2783–2792 (2010).
Canals, M. et al. Flushing submarine canyons. Nature 444, 354–357 (2006).
Harrold, C. & Lisin, S. Radio-tracking rafts of giant kelp: local production and regional transport. J. Exp. Mar. Biol. Ecol. 130, 237–251 (1989).
Baldauf, S. L. The deep roots of eukaryotes. Science 300, 1703–1706 (2003).
Zuccarello, G. C., Price, N., Verbruggen, H. & Leliaert, F. Analysis of a plastid multigene data set and the phylogenetic position of the marine macroalga Caulerpa filiformis (Chlorophyta). J. Phycol. 45, 1206–1212 (2009).
Nakada, T., Misawa, K. & Nozaki, H. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol. Phylogenet. Evol. 48, 281–291 (2008).
Saunders, G. W. & Kucera, H. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogam. Algol. 31, 487–528 (2010).
Saunders, G. W. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Phil. Trans. R. Soc. Lond. B 360, 1879–1888 (2005).
Clerissi, C. et al. Unveiling of the diversity of Prasinoviruses (Phycodnaviridae) in marine samples by using high-throughput sequencing analyses of PCR-amplified DNA polymerase and major capsid protein genes. Appl. Environ. Microbiol. 80, 3150–3160 (2014).
Pernice, M. C. et al. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 10, 945–958 (2015).
De Vargas, C. et al. List of size fractionated eukaryotic plankton community samples and associated metadata (Database W1) Pangaea https://doi.org/10.1594/PANGAEA.843017 (2015).
Lanzén, A. et al. CREST—classification resources for environmental sequence tags. PLoS ONE 7, e49334 (2012).
Cole, J. R., Konstantinidis, K., Farris, R. J. & Tiedje, J. M. in Environmental Molecular Biology (eds Liu, W.-T. & Jansson, J. K.) 1–20 (Horizon Scientific Press, 2010).
Giongo, A., Davis-Richardson, A. G., Crabb, D. B. & Triplett, E. W. TaxCollector: modifying current 16S rRNA databases for the rapid classification at six taxonomic levels. Diversity 2, 1015–1025 (2010).
Hong, S.-H., Bunge, J., Jeon, S.-O. & Epstein, S. S. Predicting microbial species richness. Proc. Natl Acad. Sci. USA 103, 117–122 (2006).
Schloss, P. D. & Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68, 686–691 (2004).
Giner, C. R. et al. Marked changes in diversity and relative activity of picoeukaryotes with depth in the global ocean. Preprint at https://www.biorxiv.org/content/10.1101/552604v1 (2019).
Krause, L. et al. Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 136, 91–101 (2008).
Huerta-Cepas, J. et al. EggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2015).
Oksanen, J. et al. vegan: Community ecology package. R package version 2.5-5 https://cran.r-project.org/web/packages/vegan/index.html (2007).
Hammer, Ř., Harper, D. A. T. & Ryan, P. D. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 4 (2001).
Acknowledgements
We thank the Tara Oceans Consortium for data availability. This research was supported by the Malaspina 2010 expedition, funded by the Spanish Ministry of Economy and Competitiveness through the Consolider-Ingenio programme to C.M.D. (reference: CSD2008-00077); CARMA, funded by the Independent Research Fund Denmark to D.K.-J. (reference: 8021-00222B); and King Abdullah University of Science and Technology’s project BAS/1/1071-01-01 to C.M.D. We thank all of the scientists and crew for support during sample collection on the Malaspina 2010 cruise, and especially E. Borrull, C. Díez-Vives, E. Lara, D. Vaqué, G. Salazar and F. Cornejo-Castillo for DNA sampling. The authors are grateful to the KAUST Supercomputing Laboratory (KSL) for the resources provided.
Author information
Authors and Affiliations
Contributions
C.M.D. and D.K.-J. conceived the research. J.M.G., S.G.A., R.L., R.M., I.A. and A.A.K. produced and curated the data. A.O., C.M.D., N.R.G. and I.A. conducted the data analysis. A.O. and C.M.D. wrote the manuscript. All co-authors contributed to improving the manuscript and approved the submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–4
Rights and permissions
About this article
Cite this article
Ortega, A., Geraldi, N.R., Alam, I. et al. Important contribution of macroalgae to oceanic carbon sequestration. Nat. Geosci. 12, 748–754 (2019). https://doi.org/10.1038/s41561-019-0421-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41561-019-0421-8
This article is cited by
-
Marine ecosystem shifts with deglacial sea-ice loss inferred from ancient DNA shotgun sequencing
Nature Communications (2023)
-
Methane emissions offset atmospheric carbon dioxide uptake in coastal macroalgae, mixed vegetation and sediment ecosystems
Nature Communications (2023)
-
Photosynthetic CO2 uptake by Ulva (Chlorophyta) as a potential contribution to global warming containment
Journal of Applied Phycology (2023)
-
A global dataset of seaweed net primary productivity
Scientific Data (2022)
-
Video survey of deep benthic macroalgae and macroalgal detritus along a glacial Arctic fjord: Kongsfjorden (Spitsbergen)
Polar Biology (2022)