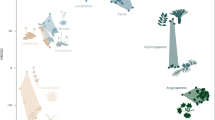
Abstract
Organismal-grade multicellularity has been achieved only in animals, plants and fungi. All three kingdoms manifest phenotypically disparate body plans but their evolution has only been considered in detail for animals. Here we tested the general relevance of hypotheses on the evolutionary assembly of animal body plans by characterizing the evolution of fungal phenotypic variety (disparity). The distribution of living fungal form is defined by four distinct morphotypes: flagellated; zygomycetous; sac-bearing; and club-bearing. The discontinuity between morphotypes is a consequence of extinction, indicating that a complete record of fungal disparity would present a more homogeneous distribution of form. Fungal disparity expands episodically through time, punctuated by a sharp increase associated with the emergence of multicellular body plans. Simulations show these temporal trends to be non-random and at least partially shaped by hierarchical contingency. These trends are decoupled from changes in gene number, genome size and taxonomic diversity. Only differences in organismal complexity, characterized as the number of traits that constitute an organism, exhibit a meaningful relationship with fungal disparity. Both animals and fungi exhibit episodic increases in disparity through time, resulting in distributions of form made discontinuous by extinction. These congruences suggest a common mode of multicellular body plan evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All original data (empirical and simulated) used in this study have been deposited at Dryad63 and are publicly available at https://doi.org/10.5061/dryad.wwpzgmsm9.
Code availability
All code used in this study has been deposited at Dryad63 and is publicly available at https://doi.org/10.5061/dryad.wwpzgmsm9.
References
Niklas, K. J. & Newman, S. A. The many roads to and from multicellularity. J. Exp. Bot. 71, 3247–3253 (2020).
Maynard Smith, J. & Szathmary, E. The Major Transitions in Evolution (W. H. Freeman Spektrum, 1995).
Grosberg, R. K. & Strathmann, R. R. The evolution of multicellularity: a minor major transition?. Annu. Rev. Ecol. Evol. Syst. 38, 621–654 (2007).
Erwin, D. H. Disparity: morphological pattern and developmental context. Palaeontology 50, 57–73 (2007).
Foote, M. The evolution of morphological diversity. Annu. Rev. Ecol. Syst. 28, 129–152 (1997).
Deline, B. et al. Evolution of metazoan morphological disparity. Proc. Natl Acad. Sci. USA 115, E8909–E8918 (2018).
Hughes, M., Gerber, S. & Wills, M. A. Clades reach highest morphological disparity early in their evolution. Proc. Natl Acad. Sci. USA 110, 13875–13879 (2013).
Kües, U., Khonsuntia, W. & Subba, S. Complex fungi. Fungal Biol. Rev. 32, 205–218 (2018).
Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438 (2011).
Li, Y. et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 31, 1653–1665.e5 (2021).
Chang, Y. et al. Genome-scale phylogenetic analyses confirm Olpidium as the closest living zoosporic fungus to the non-flagellated, terrestrial fungi. Sci. Rep. 11, 3217 (2021).
James, T. Y., Stajich, J. E., Hittinger, C. T. & Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 74, 291–313 (2020).
Naranjo-Ortiz, M. A. & Gabaldón, T. Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 94, 2101–2137 (2019).
Celio, G. J., Padamsee, M., Dentinger, B. T. M., Bauer, R. & McLaughlin, D. J. Assembling the Fungal Tree of Life: constructing the structural and biochemical database. Mycologia 98, 850–859 (2006).
Gerber, S. Use and misuse of discrete character data for morphospace and disparity analyses. Palaeontology 62, 305–319 (2019).
Smith, T. J., Puttick, M. N., O’Reilly, J. E., Pisani, D. & Donoghue, P. C. J. Phylogenetic sampling affects evolutionary patterns of morphological disparity. Palaeontology 64, 765–787 (2021).
Adl, S. M. et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019).
Berbee, M. L. et al. Genomic and fossil windows into the secret lives of the most ancient fungi. Nat. Rev. Microbiol. 18, 717–730 (2020).
Taylor, T. N., Krings, M. & Taylor, E. Fossil Fungi (Elsevier, 2015).
McShea, D. W. & Brandon, R. N. Biology’s First Law: the Tendency for Diversity and Complexity to Increase in Evolutionary Systems (Univ. of Chicago Press, 2010).
McShea, D. W. Metazoan complexity and evolution: is there a trend? Evolution 50, 477–492 (1996).
Ispolatov, I., Alekseeva, E. & Doebeli, M. Competition-driven evolution of organismal complexity. PLoS Comput. Biol. 15, e1007388 (2019).
Hobern, D. et al. Towards a global list of accepted species VI: The Catalogue of Life checklist. Org. Divers. Evol. 21, 677–690 (2021).
Bauer, R. et al. Entorrhizomycota: a new fungal phylum reveals new perspectives on the evolution of Fungi. PLoS ONE 10, e0128183 (2015).
Jones, M. D. M. et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203 (2011).
Yoshida, M., Nakayama, T. & Inouye, I. Nuclearia thermophila sp. nov. (Nucleariidae), a new nucleariid species isolated from Yunoko Lake in Nikko (Japan). Eur. J. Protistol. 45, 147–155 (2009).
Hibbett, D. S. et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111, 509–547 (2007).
Grigoriev, I. V. et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704 (2014).
Guillerme, T. & Cooper, N. Time for a rethink: time sub-sampling methods in disparity-through-time analyses. Palaeontology 61, 481–493 (2018).
Nagy, L. G., Kovács, G. M. & Krizsán, K. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biol. Rev. Camb. Philos. Soc. 93, 1778–1794 (2018).
Whittaker, R. H. New concepts of kingdoms of organisms: evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science 163, 150–160 (1969).
Naranjo-Ortiz, M. A. & Gabaldón, T. Fungal evolution: cellular, genomic and metabolic complexity. Biol. Rev. Camb. Philos. Soc. 95, 1198–1232 (2020).
Nguyen, T. A. et al. Innovation and constraint leading to complex multicellularity in the Ascomycota. Nat. Commun. 8, 14444 (2017).
Briggs, D. E., Fortey, R. A. & Wills, M. A. Morphological disparity in the Cambrian. Science 256, 1670–1673 (1992).
Gould, S. J. Wonderful Life: the Burgess Shale and the Nature of History (Hutchinson Radius, 1990).
Wan, J. et al. Decoupling of morphological disparity and taxonomic diversity during the end-Permian mass extinction. Paleobiology 47, 402–417 (2021).
Grossnickle, D. M. & Newham, E. Therian mammals experience an ecomorphological radiation during the Late Cretaceous and selective extinction at the K-Pg boundary. Proc. Biol. Sci. 283, 20160256 (2016).
Ruta, M., Angielczyk, K. D., Fröbisch, J. & Benton, M. J. Decoupling of morphological disparity and taxic diversity during the adaptive radiation of anomodont therapsids. Proc. Biol. Sci. 280, 20131071 (2013).
Bapst, D. W., Bullock, P. C., Melchin, M. J., Sheets, H. D. & Mitchell, C. E. Graptoloid diversity and disparity became decoupled during the Ordovician mass extinction. Proc. Natl Acad. Sci. USA 109, 3428–3433 (2012).
Guillerme, T. et al. Disparities in the analysis of morphological disparity. Biol. Lett. 16, 20200199 (2020).
Guillerme, T., Puttick, M. N., Marcy, A. E. & Weisbacker, V. Shifting spaces: which disparity or dissimilarity measurement best summarize occupancy in multidimensional spaces? Ecol. Evol. 10, 7261–7275 (2020).
Svardal, H., Rueffler, C. & Doebeli, M. Organismal complexity and the potential for evolutionary diversification. Evolution 68, 3248–3259 (2014).
Tenaillon, O., Silander, O. K., Uzan, J.-P. & Chao, L. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2, e217 (2007).
Valentine, J. W., Collins, A. G. & Meyer, C. P. Morphological complexity increase in metazoans. Paleobiology 20, 131–142 (1994).
Yang, J., Lusk, R. & Li, W.-H. Organismal complexity, protein complexity, and gene duplicability. Proc. Natl Acad. Sci. USA 100, 15661–15665 (2003).
Prochnik, S. E. et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223–226 (2010).
Hobern, D. et al. Towards a global list of accepted species VI: the Catalogue of Life checklist. Org. Divers. Evol. 21, 677–690 (2021).
Guillerme, T. dispRity: a modular R package for measuring disparity. Methods Ecol. Evol. 9, 1755–1763 (2018).
Tedersoo, L. et al. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 90, 135–159 (2018).
He, M.-Q. et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99, 105–367 (2019).
Beimforde, C. et al. Estimating the Phanerozoic history of the Ascomycota lineages: combining fossil and molecular data. Mol. Phylogenet. Evol. 78, 386–398 (2014).
Bollback, J. P. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88 (2006).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
O’Reilly, J. E. et al. Bayesian methods outperform parsimony but at the expense of precision in the estimation of phylogeny from discrete morphological data. Biol. Lett. 12, 20160081 (2016).
Sanderson, M. J. & Donoghue, M. J. Patterns of variation in levels of homoplasy. Evolution 43, 1781–1795 (1989).
Lloyd, G. T. Estimating morphological diversity and tempo with discrete character-taxon matrices: implementation, challenges, progress, and future directions. Biol. J. Linn. Soc. Lond. 118, 131–151 (2016).
Gower, J. C. General coefficient of similarity and some of its properties. Biometrics 27, 857–871 (1971).
Anderson, P. S. L. & Friedman, M. Using cladistic characters to predict functional variety: experiments using early gnathostomes. J. Vertebr. Paleontol. 32, 1254–1270 (2012).
Wills, M. A. in Fossils, Phylogeny, and Form: an Analytical Approach (eds Adrain, J. M. et al.) 55–144 (Springer, 2001).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Clarke, K. R. & Warwick, R. M. Change in Marine Communities: an Approach to Statistical Analysis and Interpretation (PRIMER-E Ltd, 2001).
Cailliez, F. The analytical solution of the additive constant problem. Psychometrika 48, 305–308 (1983).
Smith, T. J. & Donoghue, P. C. J. Data from: Evolution of fungal phenotypic disparity, Dryad, Dataset (2022). https://doi.org/10.5061/dryad.wwpzgmsm9
Letcher, P. M. & Powell, M. J. A taxonomic summary and revision of Rozella (Cryptomycota). IMA Fungus 9, 383–399 (2018).
James, T., Porter, T. M. & Martin, W. W. in Systematics and Evolution. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research) Vol. 7A (eds McLaughlin, D. J. & Spatafora, J. W.), 177–207 (Springer, 2014).
Seto, K., Van den Wyngaert, S., Degawa, Y. & Kagami, M. Taxonomic revision of the genus Zygorhizidium: Zygorhizidiales and Zygophlyctidales ord. nov. (Chytridiomycetes, Chytridiomycota). Fungal Syst. Evol. 5, 17–38 (2020).
Joshi, A. et al. Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 89–110 (2018).
Błaszkowski, J. et al. Dominikia bonfanteae and Glomus atlanticum, two new species in the Glomeraceae (phylum Glomeromycota) with molecular phylogenies reconstructed from two unlinked loci. Mycol. Prog. 20, 131–148 (2021).
Walther, G., Wagner, L. & Kurzai, O. Outbreaks of Mucorales and the species involved. Mycopathologia 185, 765–781 (2020).
Reynolds, N. K. et al. Phylogenetic and morphological analyses of the mycoparasitic genus Piptocephalis. Mycologia 111, 54–68 (2019).
Acknowledgements
We thank G. Storey, A. Larkin, D. Rainey, R. Wheeler and all the other Twitter users who kindly provided photographs of fungi for consideration for inclusion in this paper. We also thank P. Godoy for his thoughtful comments during the review process; the manuscript was much improved for his input. T.J.S. was funded by a Natural Environment Research Council (NERC) PhD Studentship within the GW4+ Doctoral Training Programme. P.C.J.D. was funded by the NERC (no. NE/P013678/1; part of the Biosphere Evolution, Transitions and Resilience programme, cofunded by the Natural Science Foundation of China), the Biotechnology and Biological Sciences Research Council (nos. BB/T012773/1 and BB/N000919/1), the Gordon and Betty Moore Foundation (no. GBMF9741), the John Templeton Foundation (grant no. 62220; the opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation) and the Leverhulme Trust (no. RF-2022–167).
Author information
Authors and Affiliations
Contributions
Both authors contributed to the conceptualization and design of the study, its component experiments and the interpretation of the results. T.J.S. collected the data, conducted the analyses and drafted the manuscript, to which P.C.J.D. contributed.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks P. Godoy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mean distance from centroid of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 2 Sum of variances of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 3 Average nearest neighbour Euclidean distance of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 4 Subcellular mean distance from centroid of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 5 Subcellular sum of variances of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 6 Supracellular mean distance from centroid of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 7 Supracellular sum of variances of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Extended Data Fig. 8 Supracellular average nearest neighbour Euclidean distance of 1000 bootstraps of the four morphotypes, Dikarya, and non-dikaryotic fungi.
Box plot whiskers extend to minima and maxima of data; boxes capture interquartile range and median.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and references.
Supplementary Data 1
Discrete character dataset characterizing fungal phenotypic variety.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Smith, T.J., Donoghue, P.C.J. Evolution of fungal phenotypic disparity. Nat Ecol Evol 6, 1489–1500 (2022). https://doi.org/10.1038/s41559-022-01844-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-022-01844-6
This article is cited by
-
Evolution of phenotypic disparity in the plant kingdom
Nature Plants (2023)