Abstract
The latitudinal diversity gradient (LDG) is one of the most recognized global patterns of species richness exhibited across a wide range of taxa. Numerous hypotheses have been proposed in the past two centuries to explain LDG, but rigorous tests of the drivers of LDGs have been limited by a lack of high-quality global species richness data. Here we produce a high-resolution (0.025° × 0.025°) map of local tree species richness using a global forest inventory database with individual tree information and local biophysical characteristics from ~1.3 million sample plots. We then quantify drivers of local tree species richness patterns across latitudes. Generally, annual mean temperature was a dominant predictor of tree species richness, which is most consistent with the metabolic theory of biodiversity (MTB). However, MTB underestimated LDG in the tropics, where high species richness was also moderated by topographic, soil and anthropogenic factors operating at local scales. Given that local landscape variables operate synergistically with bioclimatic factors in shaping the global LDG pattern, we suggest that MTB be extended to account for co-limitation by subordinate drivers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The global map of tree species richness is available under license CC BY 4.0, with the identifier: 10.6084/m9.figshare.17232491. This map can be downloaded in two formats. One is a geoTIFF file (S_mean_raster.tif) containing the fully geo-referenced map of tree species richness worldwide at a 0.025° × 0.025° resolution. The other is a comma-separated file (S_mean_grid.csv) with the following attributes: S is local average tree species richness per ha and x, y are centroid coordinates of all 0.025° × 0.025° pixels. The global map of co-limitation is available under license CC BY 4.0, with the identifier: 10.6084/m9.figshare.17234339. The metadata of the entire training dataframe—including the characteristics and references of all the in situ Phase I and Phase II datasets and the definitions, units and summary statistics of the environmental covariates—is available under license CC BY 4.0 with the identifier: 10.6084/m9.figshare.19733449.v1. The public version of the training dataframe, including the plot-level species richness and all the covariates, which is needed to reproduce the models and results presented here, is available at: https://doi.org/10.6084/m9.figshare.20055488. The maps and dataframe are also available on the international web research platform: Science-i (https://science-i.org/). Raw forest inventory data are commonly subject to a wide array of confidentiality clauses in regard to open access policies. Despite recent efforts to make some of these data fully open76,77, some governments and private data owners, especially those from the developing countries generally have decided to keep their data confidential. This decision is based on well-founded arguments to protect certain trees or forests (because of their large size or protected taxonomic status) from illegal logging or trespassing and to protect landowners’ privacy against the misuse of plot information such as the geographic coordinates. The sensitive information in the training dataframe, including the plot coordinates and tree-level information, will be available from the corresponding author (albeca.liang@gmail.com) upon a request via Science-i (https://science-i.org/) or GFBI (https://www.gfbinitiative.org/) and an approval from data contributors.
Code availability
All the models in this study were constructed using command line applications written in the R programming language, which processed and restructured the input data, trained the model and performed cross validation. Due to the massive amount of data, we used Purdue University’s Brown supercomputing cluster to accelerate the training process. The development of the GFBi database, tabular data cleaning, creation of species abundance matrices, evaluation of diversity determinants and geostatistical imputation were conducted in R49 (v.3.4.2) through the use of several Linux-based high-performance computing resources at Purdue University and a custom HPC interface developed using Amazon Web Services, each designed for batch processing, scalable resource distribution, embarrassingly parallel computations and/or large RAM jobs. Compute nodes with up to 1 TB of RAM and clusters of up to 64 nodes were employed in this study. Portions of the covariate preparation, mapping and quality control assessment were conducted on Windows-based operating systems with up to 128 GB of RAM. Final continental-level RF models and the R codes we developed to train the models are available under license MIT with the identifier: 10.6084/m9.figshare.17234729.
References
Sutherland, W. J. et al. Identification of 100 fundamental ecological questions. J. Ecol. 101, 58–67 (2013).
Gaston, K. J. Global patterns in biodiversity. Nature 405, 220–227 (2000).
Pickering, C. The Geographical Distribution of Animals and Plants (Little, Brown & Co., 1854).
Humboldt, A. V. & Bonpland, A. Essai sur la Géographie des Plantes Accompagné d’un Tableau Physique des Régions Équinoxiales (Levrault, Schoell & Co., 1805).
Hillebrand, H. On the generality of the latitudinal diversity gradient. Am. Nat. 163, 192–211 (2004).
Crame, J. A. Taxonomic diversity gradients through geological time. Diversity Distrib. 7, 175–189 (2001).
Gough, L. & Field, R. Latitudinal diversity gradients. eLS https://doi.org/10.1002/9780470015902.a0003233.pub2 (2007).
Willig, M. R., Kaufman, D. M. & Stevens, R. D. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol., Evolution, Syst. 34, 273–309 (2003).
Pontarp, M. et al. The latitudinal diversity gradient: novel understanding through mechanistic eco-evolutionary models. Trends Ecol. Evol. 34, 211–223 (2019).
Vellend, M. The Theory of Ecological Communities (MPB-57) Vol. 75 (Princeton Univ. Press, 2016).
Brown, J. H. Why are there so many species in the tropics? J. Biogeogr. 41, 8–22 (2014).
Currie, D. J. et al. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol. Lett. 7, 1121–1134 (2004).
Baldeck, C. A. et al. Soil resources and topography shape local tree community structure in tropical forests. Proc. R. Soc. B 280, 20122532 (2013).
Qian, H. & Ricklefs, R. E. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182 (2000).
Stein, A., Gerstner, K. & Kreft, H. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol. Lett. 17, 866–880 (2014).
Sponsel, L. E. in Encyclopedia of Biodiversity (ed. Scheiner, S.M.) 137–152 (Academic Press, 2013).
Sullivan, M. J. et al. Diversity and carbon storage across the tropical forest biome. Sci. Rep. 7, 39102 (2017).
Cazzolla Gatti, R. et al. The number of tree species on Earth. Proc. Natl Acad. Sci. USA 119, e2115329119 (2022).
Allen, A. P., Brown, J. H. & Gillooly, J. F. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science 297, 1545–1548 (2002).
Stegen, J. C., Enquist, B. J. & Ferriere, R. Advancing the metabolic theory of biodiversity. Ecol. Lett. 12, 1001–1015 (2009).
Wright, D. H. Species-energy theory: an extension of species-area theory. Oikos 41, 496–506 (1983).
Hawkins, B. A. et al. Energy, water, and broad-scale geographic patterns of species richness. Ecology 84, 3105–3117 (2003).
Staal, A., Dekker, S. C., Xu, C. & van Nes, E. H. Bistability, spatial interaction, and the distribution of tropical forests and savannas. Ecosystems 19, 1080–1091 (2016).
de L. Dantas, V., Batalha, M. A. & Pausas, J. G. Fire drives functional thresholds on the savanna–forest transition. Ecology 94, 2454–2463 (2013).
Bodart, C. et al. Continental estimates of forest cover and forest cover changes in the dry ecosystems of Africa between 1990 and 2000. J. Biogeogr. 40, 1036–1047 (2013).
Hubau, W. et al. The persistence of carbon in the African forest understory. Nat. Plants 5, 133–140 (2019).
Šímová, I. & Storch, D. The enigma of terrestrial primary productivity: measurements, models, scales and the diversity–productivity relationship. Ecography 40, 239–252 (2017).
Kreft, H. & Jetz, W. Global patterns and determinants of vascular plant diversity. Proc. Natl Acad. Sci. USA 104, 5925–5930 (2007).
Clark, D. A. et al. Net primary production in tropical forests: an evaluation and synthesis of existing field data. Ecol. Appl. 11, 371–384 (2001).
Pärtel, M., Laanisto, L. & Zobel, M. Contrasting plant productivity–diversity relationships across latitude: the role of evolutionary history. Ecology 88, 1091–1097 (2007).
Liang, J. et al. Positive biodiversity–productivity relationship predominant in global forests. Science 354, aaf8957 (2016).
Hammond, W. M. et al. Global field observations of tree die-off reveal hotter-drought fingerprint for Earth’s forests. Nat. Commun. 13, 1761 (2022).
Martens, C. et al. Large uncertainties in future biome changes in Africa call for flexible climate adaptation strategies. Glob. Change Biol. 27, 340–358 (2021).
Chapin, F. S., Bloom, A. J., Field, C. B. & Waring, R. H. Plant responses to multiple environmental factors. BioScience 37, 49–57 (1987).
Tilman, D. Resource Competition and Community Structure (MPB-17) Vol. 17 (Princeton Univ. Press, 1982).
Harpole, W. S. et al. Nutrient co-limitation of primary producer communities. Ecol. Lett. 14, 852–862 (2011).
Colwell, R. K., Rahbek, C. & Gotelli, N. J. The mid-domain effect and species richness patterns: what have we learned so far? Am. Nat.163, E1–E23 (2004).
Feng, G. et al. Species and phylogenetic endemism in angiosperm trees across the Northern Hemisphere are jointly shaped by modern climate and glacial–interglacial climate change. Glob. Ecol. Biogeogr. 28, 1393–1402 (2019).
Fraser, C. I., Nikula, R., Ruzzante, D. E. & Waters, J. M. Poleward bound: biological impacts of Southern Hemisphere glaciation. Trends Ecol. Evol. 27, 462–471 (2012).
Algar, A. C., Kerr, J. T. & Currie, D. J. A test of metabolic theory as the mechanism underlying broad-scale species-richness gradients. Glob. Ecol. Biogeogr. 16, 170–178 (2007).
Wilson, E. O. A global biodiversity map. Science 289, 2279–2279 (2000).
Castagneyrol, B. & Jactel, H. Unraveling plant–animal diversity relationships: a meta-regression analysis. Ecology 93, 2115–2124 (2012).
Steidinger, B. S. et al. Climatic controls of decomposition drive the global biogeography of forest–tree symbioses. Nature 569, 404–408 (2019).
Bongers, F. J. et al. Functional diversity effects on productivity increase with age in a forest biodiversity experiment. Nat. Ecol. Evol. 5, 1594–1603 (2021).
UN Statistical Commission. System of Environmental–Economic Accounting—Ecosystem Accounting: Final Draft (United Nations, 2021).
Green Bond Impact Report (The World Bank Treasury, 2019).
Crowther, T. et al. Mapping tree density at a global scale. Nature 525, 201–205 (2015).
Chamberlain, J. L., Prisley, S. & McGuffin, M. Understanding the relationships between American ginseng harvest and hardwood forests inventory and timber harvest to improve co-management of the forests of eastern United States. J. Sustainable For. 32, 605–624 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
Borda-de-Água, L., Hubbell, S. P. & McAllister, M. Species-area curves, diversity indices, and species abundance distributions: a multifractal analysis. Am. Nat. 159, 138–155 (2002).
Connor, E. F. & McCoy, E. D. The statistics and biology of the species-area relationship. Am. Nat. 113, 791–833 (1979).
He, F. & Legendre, P. On species-area relations. Am. Nat. 148, 719–737 (1996).
White, E. P., Ernest, S. K. M., Kerkhoff, A. J. & Enquist, B. J. Relationships between body size and abundance in ecology. Trends Ecol. Evol. 22, 323–330 (2007).
Gotelli, N. J. & Colwell, R. K. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391 (2001).
Chirici, G., Winter, S. & McRoberts, R. E. National Forest Inventories: Contributions to Forest Biodiversity Assessments Vol. 20 (Springer Science & Business Media, 2011).
Tomppo, E. et al. National Forest Inventories: Pathways for Common Reporting (Springer, 2010).
Winter, S., Böck, A. & McRoberts, R. E. Estimating tree species diversity across geographic scales. Eur. J. For. Res. 131, 441–451 (2012).
McRoberts, R. E., Winter, S., Chirici, G. & LaPoint, E. Assessing forest naturalness. For. Sci. 58, 294–309 (2012).
Release 10.3 of Desktop, ESRI ArcGIS (Environmental Systems Research Institute, 2014).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Strobl, C., Boulesteix, A.-L., Kneib, T., Augustin, T. & Zeileis, A. Conditional variable importance for random forests. BMC Bioinf. 9, 307 (2008).
Olson, D. M. & Dinerstein, E. The global 200: priority ecoregions for global conservation. Ann. Mo. Bot. Gard. 89, 199–224 (2002).
Hyndman, R. J. & Koehler, A. B. Another look at measures of forecast accuracy. Int. J. Forecast. 22, 679–688 (2006).
Legendre, P. Spatial autocorrelation: trouble or new paradigm? Ecology 74, 1659–1673 (1993).
Husch, B., Beers, T. W. & Kershaw Jr, J. A. Forest Mensuration 4th edn (John Wiley & Sons, 2003).
Farrar, D. E. & Glauber, R. R. Multicollinearity in regression analysis: the problem revisited. Rev. Econ. Stat. 49, 92–107 (1967).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S-PLUS (Springer Science & Business Media, 2013).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (Association for Computing Machinery, 2016).
Friedman, J. H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 29, 1189–1232 (2001).
Hansen, M. C. et al. High-resolution global maps of 21st-century forest cover change. Science 342, 850–853 (2013).
Global Forest Resources Assessment 2015—How are the World’s Forests Changing? (Food and Agriculture Organization of the United Nations, 2015).
Bastin, J.-F. et al. The extent of forest in dryland biomes. Science 356, 635–638 (2017).
Peres-Neto, P. R., Legendre, P., Dray, S. & Borcard, D. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87, 2614–2625 (2006).
Peres-Neto, P. R. & Legendre, P. Estimating and controlling for spatial structure in the study of ecological communities. Glob. Ecol. Biogeogr. 19, 174–184 (2010).
Saltelli, A. et al. Global Sensitivity Analysis: the Primer (John Wiley & Sons, 2008).
Liang, J. & Gamarra, J. G. P. The importance of sharing global forest data in a world of crises. Sci. Data 7, 424 (2020).
Towards Open and Transparent Forest Data for Climate Action—Experiences and Lessons Learned (United Nations Food and Agriculture Organization, 2022).
Acknowledgements
The team collaboration and manuscript development are supported by the web-based team science platform: science-i.org, with the project number 202205GFB2. We thank the following initiatives, agencies, teams and individuals for data collection and other technical support: the Global Forest Biodiversity Initiative (GFBI) for establishing the data standards and collaborative framework; United States Department of Agriculture, Forest Service, Forest Inventory and Analysis (FIA) Program; University of Alaska Fairbanks; the SODEFOR, Ivory Coast; University Félix Houphouët-Boigny (UFHB, Ivory Coast); the Queensland Herbarium and past Queensland Government Forestry and Natural Resource Management departments and staff for data collection for over seven decades; and the National Forestry Commission of Mexico (CONAFOR). We thank M. Baker (Carbon Tanzania), together with a team of field assistants (Valentine and Lawrence); all persons who made the Third Spanish Forest Inventory possible, especially the main coordinator, J. A. Villanueva (IFN3); the French National Forest Inventory (NFI campaigns (raw data 2005 and following annual surveys, were downloaded by GFBI at https://inventaire-forestier.ign.fr/spip.php?rubrique159; site accessed on 1 January 2015)); the Italian Forest Inventory (NFI campaigns raw data 2005 and following surveys were downloaded by GFBI at https://inventarioforestale.org/; site accessed on 27 April 2019); Swiss National Forest Inventory, Swiss Federal Institute for Forest, Snow and Landscape Research WSL and Federal Office for the Environment FOEN, Switzerland; the Swedish NFI, Department of Forest Resource Management, Swedish University of Agricultural Sciences SLU; the National Research Foundation (NRF) of South Africa (89967 and 109244) and the South African Research Chair Initiative; the Danish National Forestry, Department of Geosciences and Natural Resource Management, UCPH; Coordination for the Improvement of Higher Education Personnel of Brazil (CAPES, grant number 88881.064976/2014-01); R. Ávila and S. van Tuylen, Instituto Nacional de Bosques (INAB), Guatemala, for facilitating Guatemalan data; the National Focal Center for Forest condition monitoring of Serbia (NFC), Institute of Forestry, Belgrade, Serbia; the Thünen Institute of Forest Ecosystems (Germany) for providing National Forest Inventory data; the FAO and the United Nations High Commissioner for Refugees (UNHCR) for undertaking the SAFE (Safe Access to Fuel and Energy) and CBIT-Forest projects; and the Amazon Forest Inventory Network (RAINFOR), the African Tropical Rainforest Observation Network (AfriTRON) and the ForestPlots.net initiative for their contributions from Amazonian and African forests. The Natural Forest plot data collected between January 2009 and March 2014 by the LUCAS programme for the New Zealand Ministry for the Environment are provided by the New Zealand National Vegetation Survey Databank https://nvs.landcareresearch.co.nz/. We thank the International Boreal Forest Research Association (IBFRA); the Forestry Corporation of New South Wales, Australia; the National Forest Directory of the Ministry of Environment and Sustainable Development of the Argentine Republic (MAyDS) for the plot data of the Second National Forest Inventory (INBN2); the National Forestry Authority and Ministry of Water and Environment of Uganda for their National Biomass Survey (NBS) dataset; and the Sabah Biodiversity Council and the staff from Sabah Forest Research Centre. All TEAM data are provided by the Tropical Ecology Assessment and Monitoring (TEAM) Network, a collaboration between Conservation International, the Missouri Botanical Garden, the Smithsonian Institution and the Wildlife Conservation Society, and partially funded by these institutions, the Gordon and Betty Moore Foundation and other donors, with thanks to all current and previous TEAM site manager and other collaborators that helped collect data. We thank the people of the Redidoti, Pierrekondre and Cassipora village who were instrumental in assisting with the collection of data and sharing local knowledge of their forest and the dedicated members of the field crew of Kabo 2012 census. We are also thankful to FAPESC, SFB, FAO and IMA/SC for supporting the IFFSC. This research was supported in part through computational resources provided by Information Technology at Purdue, West Lafayette, Indiana.This work is supported in part by the NASA grant number 12000401 ‘Multi-sensor biodiversity framework developed from bioacoustic and space based sensor platforms’ (J. Liang, B.P.); the USDA National Institute of Food and Agriculture McIntire Stennis projects 1017711 (J. Liang) and 1016676 (M.Z.); the US National Science Foundation Biological Integration Institutes grant NSF‐DBI‐2021898 (P.B.R.); the funding by H2020 VERIFY (contract 776810) and H2020 Resonate (contract 101000574) (G.-J.N.); the TEAM project in Uganda supported by the Moore foundation and Buffett Foundation through Conservation International (CI) and Wildlife Conservation Society (WCS); the Danish Council for Independent Research | Natural Sciences (TREECHANGE, grant 6108-00078B) and VILLUM FONDEN grant number 16549 (J.-C.S.); the Natural Environment Research Council of the UK (NERC) project NE/T011084/1 awarded to J.A.-G. and NE/ S011811/1; ERC Advanced Grant 291585 (‘T-FORCES’) and a Royal Society-Wolfson Research Merit Award (O.L.P.); RAINFOR plots supported by the Gordon and Betty Moore Foundation and the UK Natural Environment Research Council, notably NERC Consortium Grants ‘AMAZONICA’ (NE/F005806/1), ‘TROBIT’ (NE/D005590/1) and ‘BIO-RED’ (NE/N012542/1); CIFOR’s Global Comparative Study on REDD+ funded by the Norwegian Agency for Development Cooperation, the Australian Department of Foreign Affairs and Trade, the European Union, the International Climate Initiative (IKI) of the German Federal Ministry for the Environment, Nature Conservation, Building and Nuclear Safety and the CGIAR Research Program on Forests, Trees and Agroforestry (CRP-FTA) and donors to the CGIAR Fund; AfriTRON network plots funded by the local communities and NERC, ERC, European Union, Royal Society and Leverhume Trust; a grant from the Royal Society and the Natural Environment Research Council, UK (S.L.L.); National Science Foundation CIF21 DIBBs: EI: number 1724728 (A.C.C.); National Natural Science Foundation of China (31800374) and Shandong Provincial Natural Science Foundation (ZR2019BC083) (H.L.). UK NERC Independent Research Fellowship (grant code: NE/S01537X/1) (T.J.); a Serra-Húnter Fellowship provided by the Government of Catalonia (Spain) (S.d.-M.); the Brazilian National Council for Scientific and Technological Development (CNPq, grant 442640/2018-8, CNPq/Prevfogo-Ibama number 33/2018) (C.A.S.); a grant from the Franklinia Foundation (D.A.C.); Russian Science Foundation project number 19-77-300-12 (R.V.); the Takenaka Scholarship Foundation (A.O.A.); the German Research Foundation (DFG), grant number Am 149/16-4 (C.A.); the Romania National Council for Higher Education Funding, CNFIS, project number CNFIS-FDI-2022-0259 (O.B.); Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-05109 and STPGP506284) and the Canadian Foundation for Innovation (36014) (H.Y.H.C.); the project SustES—Adaptation strategies for sustainable ecosystem services and food security under adverse environmental conditions (CZ.02.1.01/0.0/0.0/16_019/0000797) (E.C.); Consejo de Ciencia y Tecnología del estado de Durango (2019-01-155) (J.J.C.-R.); Science and Engineering Research Board (SERB), New Delhi, Government of India (file number PDF/2015/000447)—‘Assessing the carbon sequestration potential of different forest types in Central India in response to climate change’ (J.A.D.); Investissement d’avenir grant of the ANR (CEBA: ANR-10-LABEX-0025) (G.D.); National Foundation for Science & Technology Development of Vietnam, 106-NN.06-2013.01 (T.V.D.); Queensland government, Department of Environment and Science (T.J.E.); a Czech Science Foundation Standard grant (19-14620S) (T.M.F.); European Union Seventh Framework Program (FP7/2007–2013) under grant agreement number 265171 (L. Finer, M. Pollastrini, F. Selvi); grants from the Swedish National Forest Inventory, Swedish University of Agricultural Sciences (J.F.); CNPq productivity grant number 311303/2020-0 (A.L.d.G.); DFG grant HE 2719/11-1,2,3; HE 2719/14-1 (A. Hemp); European Union’s Horizon Europe research project OpenEarthMonitor grant number 101059548, CGIAR Fund INIT-32-MItigation and Transformation Initiative for GHG reductions of Agrifood systems RelaTed Emissions (MITIGATE+) (M.H.); General Directorate of the State Forests, Poland (1/07; OR-2717/3/11; OR.271.3.3.2017) and the National Centre for Research and Development, Poland (BIOSTRATEG1/267755/4/NCBR/2015) (A.M.J.); Czech Science Foundation 18-10781 S (S.J.); Danish of Ministry of Environment, the Danish Environmental Protection Agency, Integrated Forest Monitoring Program—NFI (V.K.J.); State of São Paulo Research Foundation/FAPESP as part of the BIOTA/FAPESP Program Project Functional Gradient-PELD/BIOTA-ECOFOR 2003/12595-7 & 2012/51872-5 (C.A.J.); Danish Council for Independent Research—social sciences—grant DFF 6109–00296 (G.A.K.); Russian Science Foundation project 21-46-07002 for the plot data collected in the Krasnoyarsk region (V.K.); BOLFOR (D.K.K.); Department of Biotechnology, New Delhi, Government of India (grant number BT/PR7928/NDB/52/9/2006, dated 29 September 2006) (M.L.K.); grant from Kenya Coastal Development Project (KCDP), which was funded by World Bank (J.N.K.); Korea Forest Service (2018113A00-1820-BB01, 2013069A00-1819-AA03, and 2020185D10-2022-AA02) and Seoul National University Big Data Institute through the Data Science Research Project 2016 (H.S.K.); the Brazilian National Council for Scientific and Technological Development (CNPq, grant 442640/2018-8, CNPq/Prevfogo-Ibama number 33/2018) (C.K.); CSIR, New Delhi, government of India (grant number 38(1318)12/EMR-II, dated: 3 April 2012) (S.K.); Department of Biotechnology, New Delhi, government of India (grant number BT/ PR12899/ NDB/39/506/2015 dated 20 June 2017) (A.K.); Coordination for the Improvement of Higher Education Personnel (CAPES) #88887.463733/2019-00 (R.V.L.); National Natural Science Foundation of China (31800374) (H.L.); project of CEPF RAS ‘Methodological approaches to assessing the structural organization and functioning of forest ecosystems’ (AAAA-A18-118052590019-7) funded by the Ministry of Science and Higher Education of Russia (N.V.L.); Leverhulme Trust grant to Andrew Balmford, Simon Lewis and Jon Lovett (A.R.M.); Russian Science Foundation, project 19-77-30015 for European Russia data processing (O.M.); grant from Kenya Coastal Development Project (KCDP), which was funded by World Bank (M.T.E.M.); the National Centre for Research and Development, Poland (BIOSTRATEG1/267755/4/NCBR/2015) (S.M.); the Secretariat for Universities and of the Ministry of Business and Knowledge of the Government of Catalonia and the European Social Fund (A. Morera); Queensland government, Department of Environment and Science (V.J.N.); Pinnacle Group Cameroon PLC (L.N.N.); Queensland government, Department of Environment and Science (M.R.N.); the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-05201) (A.P.); the Russian Foundation for Basic Research, project number 20-05-00540 (E.I.P.); European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement number 778322 (H.P.); Science and Engineering Research Board, New Delhi, government of India (grant number YSS/2015/000479, dated 12 January 2016) (P.S.); the Chilean Government research grants Fondecyt number 1191816 and FONDEF number ID19 10421 (C.S.-E.); the Deutsche Forschungsgemeinschaft (DFG) Priority Program 1374 Biodiversity Exploratories (P.S.); European Space Agency projects IFBN (4000114425/15/NL/FF/gp) and CCI Biomass (4000123662/18/I-NB) (D. Schepaschenko); FunDivEUROPE, European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement number 265171 (M.S.-L.); APVV 20-0168 from the Slovak Research and Development Agency (V.S.); Manchester Metropolitan University’s Environmental Science Research Centre (G.S.); the project ‘LIFE+ ForBioSensing PL Comprehensive monitoring of stand dynamics in Białowieża Forest supported with remote sensing techniques’ which is co-funded by the EU Life Plus programme (contract number LIFE13 ENV/PL/000048) and the National Fund for Environmental Protection and Water Management in Poland (contract number 485/2014/WN10/OP-NM-LF/D) (K.J.S.); Global Challenges Research Fund (QR allocation, MMU) (M.J.P.S.); Czech Science Foundation project 21-27454S (M.S.); the Russian Foundation for Basic Research, project number 20-05-00540 (N. Tchebakova); Botanical Research Fund, Coalbourn Trust, Bentham Moxon Trust, Emily Holmes scholarship (L.A.T.); the programmes of the current scientific research of the Botanical Garden of the Ural Branch of Russian Academy of Sciences (V.A.U.); FCT—Portuguese Foundation for Science and Technology—Project UIDB/04033/2020. Inventário Florestal Nacional—ICNF (H. Viana); Grant from Kenya Coastal Development Project (KCDP), which was funded by World Bank (C.W.); grants from the Swedish National Forest Inventory, Swedish University of Agricultural Sciences (B.W.); ATTO project (grant number MCTI-FINEP 1759/10 and BMBF 01LB1001A, 01LK1602F) (F.W.); ReVaTene/PReSeD-CI 2 is funded by the Education and Research Ministry of Côte d’Ivoire, as part of the Debt Reduction-Development Contracts (C2Ds) managed by IRD (I.C.Z.-B.); the National Research Foundation of South Africa (NRF, grant 89967) (C.H.). The Tropical Plant Exploration Group 70 1 ha plots in Continental Cameroon Mountains are supported by Rufford Small Grant Foundation, UK and 4 ha in Sierra Leone are supported by the Global Challenge Research Fund through Manchester Metropolitan University, UK; the National Geographic Explorer Grant, NGS-53344R-18 (A.C.-S.); University of KwaZulu-Natal Research Office grant (M.J.L.); Universidad Nacional Autónoma de México, Dirección General de Asuntos de Personal Académico, Grant PAPIIT IN-217620 (J.A.M.). Czech Science Foundation project 21-24186M (R.T., S. Delabye). Czech Science Foundation project 20-05840Y, the Czech Ministry of Education, Youth and Sports (LTAUSA19137) and the long-term research development project of the Czech Academy of Sciences no. RVO 67985939 (J.A.). The American Society of Primatologists, the Duke University Graduate School, the L.S.B. Leakey Foundation, the National Science Foundation (grant number 0452995) and the Wenner-Gren Foundation for Anthropological Research (grant number 7330) (M.B.). Research grants from Conselho Nacional de Desenvolvimento Científico e Tecnologico (CNPq, Brazil) (309764/2019; 311303/2020) (A.C.V., A.L.G.). The Project of Sanya Yazhou Bay Science and Technology City (grant number CKJ-JYRC-2022-83) (H.-F.W.). The Ugandan NBS was supported with funds from the Forest Carbon Partnership Facility (FCPF), the Austrian Development Agency (ADC) and FAO. FAO’s UN-REDD Program, together with the project on ‘Native Forests and Community’ Loan BIRF number 8493-AR UNDP ARG/15/004 and the National Program for the Protection of Native Forests under UNDP funded Argentina’s INBN2.
Author information
Authors and Affiliations
Contributions
Conceptualization: J. Liang and C.H. Methodology: J. Liang, C.H., J.G.P.G. and N. Picard. Data coordination: J. Liang, M.Z., S.d.-M., T.W.C., G.-J.N., P.B.R., F. Slik, K.v.G., J.G.P.G. and N. Picard. Writing, revision and editing: all.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Jennifer Baltzer and Chengjin Chu for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
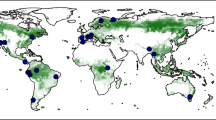
Extended Data Fig. 1 Distribution of sample plots in the global forest biodiversity individual-based database (GFBi).
(A) GFBi data consists of in situ measurements of approximately 1,255,444 forest sample plots (Phase I sample plots, blue). Together with an independent and complementary dataset of 22,131 forest sample plots (Phase II sample plots, yellow). (B) GFBi sample plots cover 424 ecoregions across the world, with the proportion of sample plots by ecoregion ranging from 0.001% to 19.333%. Phase I sample plots cover 394 ecoregions across the world, and Phase II sample plots cover an additional 30 ecoregions in Africa, South America, Southeast Asia, Mexico, India, and Japan. Together, our ground-based forest sample plots cover 424 of 435 forested ecoregions across the world.
Extended Data Fig. 2 Examination of estimated tree species richness using three cross-validation approaches.
(Left column) In randomized cross-validation (RCV), three imputation models—random forests (RF, top), multiple regression with ordinary least squares (OLS, centre), and XGBoost (XGB, bottom)—were trained for each continent with a random subsample comprising 90% of the training data from that continent; the remaining 10% of the training data were used as the testing set. This process was repeated 20 times with sample replacement to examine the accuracy of estimated tree species richness values for each sample plot. (Centre column) In spatial cross-validation (SCV), all sample data from an ecoregion were reserved for testing the three imputation models, trained with the remaining samples from the same continent. This process was repeated until all the forested ecoregions across the world had been tested. (Right column) For post-sample validation (PSV), we collated a new sample dataset from 22,131 forest sample plots (Phase II data), and used Phase II data as the testing set to evaluate the accuracy of the predictive models that were trained for each continent with the Phase I data. Scatter plots show observed (vertical axis) vs. predicted (horizontal axis) values of tree species richness per hectare, from which we calculated mean absolute error (MAE), root-mean-squared error (RMSE), and the coefficient of determination (R2) of the trendlines (red) between the predicted and observed values.
Extended Data Fig. 3 Partial dependence of tree species richness on each predictor variable.
We drew a partial dependence plot (with 95% confidential interval in shaded bands) for each of the 47 explanatory variables in five categories, that is, bioclimatic (green), vegetation and survey (orange), topographic (blue), anthropogenic (pink), and soil (lime). The final panel in the lower right corner is a 2D partial dependence plot showing how tree species richness changes by annual mean temperature and total annual precipitation, while all the other predictors remained constant at their sample mean. Curves on top of all partial dependence plots depict the density of sample data for each explanatory variable. See Supplementary Table 1 for the units and a detailed description of the explanatory variables.
Extended Data Fig. 4 Analyses of the latitudinal gradients in the residuals from three nested models.
For each model, we show a scatter plot of absolute residual values (blue circles) and a column plot of root-mean-squared error (RMSE) calculated from the training dataset, with 2° bins and vertical axis in reverse order (black columns). Model (A) consisted of all 47 explanatory variables plus latitude. Model (B) contained all the explanatory variables in (A) except for the 21 bioclimatic variables. Model (C) as the base model only contained a zero constant. RMSE values in the upper-right corner of each panel represent the total RMSE, with p-values calculated from the one-sided F-test (see Variance Partitioning for details). p(A) stands for the p-value of all 47 explanatory variables plus latitude, and p(B) stands for the p-value of 21 bioclimatic variables.
Extended Data Fig. 5 Hyper-parameter selection for the random forests model.
Using 10 bootstrapping iterations on random training sets consisting of 1000 random sample for each continent, we calculated the sensitivity of the root-mean-squared error (RMSE) to (left) the number of trees to grow and (right) the number of variables randomly sampled as candidates at each split in the random forests model. As RMSE converged at 50 trees and 10 variables, we selected them as optimal hyper-parameters for the random forests model. Black lines represent the mean of all the iterations (with red bands showing the standard error of the mean), and blues lines represent each iteration.
Extended Data Fig. 6 Hyper-parameter selection for the XGBoost model.
Using 20 bootstrapping iterations on random training sets consisting of 90% of sample for each continent, we calculated the sensitivity of the root-mean-squared error (RMSE) of the testing sets (consisting of the remaining 10% of sample) to (left) the maximum number of boosting iterations (that is number of rounds), and (right) the maximum depth of a tree for the XGBoost model. As RMSE converged at 50 rounds and 20 depth, we selected them as optimal hyper-parameters for the XGBoost model. The box plot represents the 25th and 75th percentiles (bounds of box), median (centre line), and the maximum and minimum (upper and lower whiskers) of the RMSE values for each level of tree depth.
Extended Data Fig. 7 Co-limiting environmental drivers of tree species richness per hectare in forested areas worldwide.
Driver dominance was derived for each pixel from four driver categories (that is bioclimatic, topographic, anthropogenic, and soil), as well as ‘co-limitation’ which represents a lack of clear dominance among the four categories above. All map layers are displayed at a 0.025° pixel with an equirectangular projection (Plate–Carrée). See Fig. 5 for a map of all categories overlaid on top of each other.
Supplementary information
Rights and permissions
About this article
Cite this article
Liang, J., Gamarra, J.G.P., Picard, N. et al. Co-limitation towards lower latitudes shapes global forest diversity gradients. Nat Ecol Evol 6, 1423–1437 (2022). https://doi.org/10.1038/s41559-022-01831-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-022-01831-x
This article is cited by
-
How important is passive sampling to explain species-area relationships? A global synthesis
Landscape Ecology (2024)
-
Advancing forest inventorying and monitoring
Annals of Forest Science (2024)
-
Spatial database of planted forests in East Asia
Scientific Data (2023)
-
Soundscapes and deep learning enable tracking biodiversity recovery in tropical forests
Nature Communications (2023)
-
Mapping density, diversity and species-richness of the Amazon tree flora
Communications Biology (2023)