Abstract
Deleterious mutations are generally considered to be irrelevant for morphological evolution. However, they could be compensated by conditionally beneficial mutations, thereby providing access to new adaptive paths. Here we use high-dimensional phenotyping of laboratory-evolved budding yeast lineages to demonstrate that new cellular morphologies emerge exceptionally rapidly as a by-product of gene loss and subsequent compensatory evolution. Unexpectedly, the capacities for invasive growth, multicellular aggregation and biofilm formation also spontaneously evolve in response to gene loss. These multicellular phenotypes can be achieved by diverse mutational routes and without reactivating the canonical regulatory pathways. These ecologically and clinically relevant traits originate as pleiotropic side effects of compensatory evolution and have no obvious utility in the laboratory environment. The extent of morphological diversity in the evolved lineages is comparable to that of natural yeast isolates with diverse genetic backgrounds and lifestyles. Finally, we show that both the initial gene loss and subsequent compensatory mutations contribute to new morphologies, with their synergistic effects underlying specific morphological changes. We conclude that compensatory evolution is a previously unrecognized source of morphological diversity and phenotypic novelties.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the main text, Methods or the Supplementary Information. A multi-page pdf containing the investigation of ploidy level of yeast strains is available at https://figshare.com/s/cc55743a3c97d927db59. High-resolution image of Extended Data Fig. 4 can be found at https://figshare.com/s/a5f1571eb8cc5bada89b.
Code availability
Scripts used in the analysis of microscopic images are available at https://github.com/pappb/Farkas-et-al-Compensatory-evolution. The MATLAB code used in the image analysis of invasive growth is available at https://github.com/csmolnar/invasivegrowth.
References
Covert, A. W., Lenski, R. E., Wilke, C. O. & Ofria, C. Experiments on the role of deleterious mutations as stepping stones in adaptive evolution. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1313424110 (2013).
Albalat, R. & Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016).
Lang, G. I. et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013).
Qian, W., Ma, D., Xiao, C., Wang, Z. & Zhang, J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2, 1399–1410 (2012).
Doniger, S. W. et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 4, e1000183 (2008).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Sharma, V. et al. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 9, 1215 (2018).
Szamecz, B. et al. The genomic landscape of compensatory evolution. PLoS Biol. 12, e1001935 (2014).
LaBar, T., Phoebe Hsieh, Y.-Y., Fumasoni, M. & Murray, A. W. Evolutionary repair experiments as a window to the molecular diversity of life. Curr. Biol. 30, R565–R574 (2020).
Wagner, A. Robustness, evolvability, and neutrality. FEBS Lett. 579, 1772–1778 (2005).
Lynch, M. The evolution of genetic networks by non-adaptive processes. Nat. Rev. Genet. 8, 803–813 (2007).
Ivankov, D. N., Finkelstein, A. V. & Kondrashov, F. A. A structural perspective of compensatory evolution. Curr. Opin. Struct. Biol. 26, 104–112 (2014).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010).
Wittkopp, P. J., Haerum, B. K. & Clark, A. G. Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 (2004).
Connallon, T., Camus, M. F., Morrow, E. H. & Dowling, D. K. Coadaptation of mitochondrial and nuclear genes, and the cost of mother’s curse. Proc. R. Soc. B 285, 20172257 (2018).
Galardini, M. et al. The impact of the genetic background on gene deletion phenotypes in Saccharomyces cerevisiae. Mol. Syst. Biol. 15, e8831 (2019).
Blank, D., Wolf, L., Ackermann, M. & Silander, O. K. The predictability of molecular evolution during functional innovation. Proc. Natl Acad. Sci. USA 111, 3044–3049 (2014).
McCloskey, D. et al. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 9, 3796 (2018).
Rojas Echenique, J. I., Kryazhimskiy, S., Nguyen Ba, A. N. & Desai, M. M. Modular epistasis and the compensatory evolution of gene deletion mutants. PLoS Genet. 15, e1007958 (2019).
Ohya, Y. et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc. Natl Acad. Sci. USA 102, 19015–19020 (2005).
Bauer, C. R., Li, S. & Siegal, M. L. Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol. Syst. Biol. 11, 773 (2015).
Spor, A., Wang, S., Dillmann, C., Vienne, Dde & Sicard, D. “Ant” and “grasshopper” life-history strategies in Saccharomyces cerevisiae. PLoS ONE 3, e1579 (2008).
Turner, J. J., Ewald, J. C. & Skotheim, J. M. Cell size control in yeast. Curr. Biol. 22, R350–R359 (2012).
Yvert, G. et al. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Syst. Biol. 7, 54 (2013).
Okada, H., Ohnuki, S. & Ohya, Y. Quantification of cell, actin, and nuclear DNA morphology with high-throughput microscopy and CalMorph. Cold Spring Harb. Protoc. 4, 408–412 (2015).
Suzuki, G. et al. Global study of holistic morphological effectors in the budding yeast Saccharomyces cerevisiae. BMC Genom. 19, 149 (2018).
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
Peter, J. et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018).
Watanabe, M., Watanabe, D., Nogami, S., Morishita, S. & Ohya, Y. Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr. Genet. 55, 365–380 (2009).
Gimeno, C. J., Ljungdahl, P. O., Styles, C. A. & Fink, G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077–1090 (1992).
Roberts, R. L. & Fink, G. R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8, 2974–2985 (1994).
Madhani, H. D. & Fink, G. R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 8, 348–353 (1998).
Cullen, P. J. & Sprague, G. F. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl Acad. Sci. USA 97, 13619–13624 (2000).
Reynolds, T. B. & Fink, G. R. Bakers’ yeast, a model for fungal biofilm formation. Science 291, 878–881 (2001).
Soares, E. V. Flocculation in Saccharomyces cerevisiae: a review. J. Appl. Microbiol. 110, 1–18 (2011).
Kuzdzal-Fick, J. J., Chen, L. & Balázsi, G. Disadvantages and benefits of evolved unicellularity versus multicellularity in budding yeast. Ecol. Evol. 9, 8509–8523 (2019).
Desai, J. V., Mitchell, A. P. & Andes, D. R. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 4, a019729 (2014).
Hope, E. A. & Dunham, M. J. Ploidy-regulated variation in biofilm-related phenotypes in natural isolates of Saccharomyces cerevisiae. G3 4, 1773–1786 (2014).
Liu, H., Styles, C. A. & Fink, G. R. Saccharomyces cerevisiae S288c has a mutation in Flo8, a gene required for filamentous growth. Genetics 144, 967–978 (1996).
Lo, W.-S. & Dranginis, A. M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9, 161–171 (1998).
Vermulst, M. et al. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat. Commun. 6, 8065 (2015).
Chen, X. et al. Whi2 is a conserved negative regulator of TORC1 in response to low amino acids. PLoS Genet. 14, e1007592 (2018).
Hardwick, K. G. The spindle checkpoint. Trends Genet. 14, 1–4 (1998).
Lew, D. J. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15, 648–653 (2003).
Helsen, J. et al. Gene loss predictably drives evolutionary adaptation. Mol. Biol. Evol. 37, 2989–3002 (2020).
Tan, Z. et al. Aneuploidy underlies a multicellular phenotypic switch. Proc. Natl Acad. Sci. USA 110, 12367–12372 (2013).
Ho, W.-C., Ohya, Y. & Zhang, J. Testing the neutral hypothesis of phenotypic evolution. Proc. Natl Acad. Sci. USA 114, 12219–12224 (2017).
Pavlicev, M. & Wagner, G. P. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol. Evol. 27, 316–322 (2012).
Steenwyk, J. L. et al. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biol. 17, e3000255 (2019).
Parts, L. Natural variants suppress mutations in hundreds of essential genes. Mol. Syst. Biol. 17, e10138 (2021).
Orr, H. A. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52, 935–949 (1998).
Goldschmidt, R. The Material Basis of Evolution (Yale Univ. Press, 1940).
Warringer, J., Ericson, E., Fernandez, L., Nerman, O. & Blomberg, A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl Acad. Sci. USA 100, 15724–15729 (2003).
Warringer, J. & Blomberg, A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20, 53–67 (2003).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019).
Acknowledgements
The FRE-LacZ plasmid (YEpU-FTyZ) was a kind gift from J. Thorner. We thank Z. Bódi for informal discussions, K. Ambrus for her general technical assistance, E. Kotogány for her help in the flow-cytometry measurements and I. Kelemen-Valkony for her help in laser scanning confocal microscopy. Funding and grant sources are as follows: ‘Lendület’ program of the Hungarian Academy of Sciences LP2009-013/2012 (B.P.); ‘Lendület’ program of the Hungarian Academy of Sciences LP-2017-10/2020 (C.P.); LENDULET-BIOMAG grant 2018-342 (P.H.); Wellcome Trust WT 098016/Z/11/Z (B.P.); National Laboratory of Biotechnology grant NKFIH-871-3/2020 (C.P.); the European Research Council H2020-ERC-2014-CoG 648364- Resistance Evolution (C.P.); National Research, Development and Innovation Office Élvonal Program KKP 126506 (C.P.); National Research, Development and Innovation Office Élvonal Program KKP 129814 (B.P.); Economic Development and Innovation Operational Programme: European Regional Development Funds GINOP-2.3.2-15-2016-00006 (P.H.); Economic Development and Innovation Operational Programme: European Regional Development Funds GINOP-2.3.2-15-2016-00037 (P.H.); Economic Development and Innovation Operational Programme: European Regional Development Funds GINOP-2.3.2-15-2016-00014 (C.P., B.P.); Economic Development and Innovation Operational Programme: European Regional Development Funds GINOP-2.3.2-15-2016-00020 (C.P.); Economic Development and Innovation Operational Programme: European Regional Development Funds GINOP-2.3.2-15-2016-00026 (B.P., P.H.); the European Union’s Horizon 2020 research and innovation program grant number 739593 (B.P., F.A.); COMPASS-ERA PerMed H2020 (P.H.); CZI Deep Visual Proteomics (P.H.); H2020-DiscovAir (P.H.); ELKH-Excellence grant (P.H.); Hungarian Academy of Sciences Postdoctoral Fellowship Program Postdoc2014-85 (K.K.); National Research, Development and Innovation Office FK 128775 (Z.F.); National Research, Development and Innovation Office FK 128916 (D.K.); Janos Bolyai Research Fellowship from the Hungarian Academy of Sciences BO/779/20 (Z.F.); New National Excellence Program of the Ministry of Human Capacities Bolyai+, UNKP-20-5-SZTE-646 (Z.F.); and New National Excellence Program of the Ministry of Human Capacities Bolyai+, UNKP-21-5-SZTE-562 (Z.F.).
Author information
Authors and Affiliations
Contributions
Conceptualization: B.P. and C.P. Methodology: Z.F., K.K., Z.S., D.K., G.F., F.B., F.A., C.M. and P.H. Investigation: Z.F., K.K., Z.S., G.F. and C.M. Visualization: Z.F., K.K., Z.S. and G.F. Funding acquisition: B.P., C.P. and P.H. Supervision: B.P. and C.P. Writing—original draft: B.P., C.P., Z.F., K.K. and Z.S. Writing—review and editing: B.P., C.P., Z.F., K.K. and Z.S.
Corresponding authors
Ethics declarations
Competing interests
Authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Yoshikazu Ohya, Alys Cheatle Jarvela and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
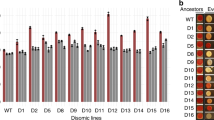
Extended Data Fig. 1 Fitness distribution of the investigated strains.
The barplot shows the distribution of relative fitness of initial knock-out mutant strains (i.e. ancestors, red) and the compensated strains (blue). Data from our previous study8 is re-plotted here. Relative fitness was estimated by growth rate in liquid medium relative to the wild-type.
Extended Data Fig. 2 Results of principal component analysis on single-cell morphology.
(a) Cumulative variance of all single-cell morphological traits explained by the first 8 principal components in a principal component analysis (PCA). Note that PCA was performed on all genotypes, including wild-type and gene deletion ancestors. (bc) Contribution of specific morphological traits to the first 8 principal components. Panels (b) and (c) show the results of separate PCAs carried out for all strains and the subset of novel compensated strains (including the WT), respectively. Colors of the bars indicate the sign of the effects of specific traits on the given principal component (loading). Only traits providing the 8 largest contributions to the principal components are shown. Note that the traits contributing to PC1 to PC8 in panels (b) and (c) show substantial overlap with each other.
Extended Data Fig. 3 Evolution of five representative morphological traits in compensated strains with the most extreme trait values.
The boxplots display the trait values in the compensated strains and corresponding ancestors compared to that of the wild-type (based on n = 3 or n = 4 biological replicates each). Traits are representative traits of PCs 3-7, shown in the order of PCs (Fig. 1a). Note that for each trait, a subset of compensated strains displaying the most extreme trait values are displayed. The corresponding CalMorph traits are D103_C, D148_A, C118_A1B, C118_C, and D182_A respectively. Dashed lines indicate the range of the wild-type trait values (average ± 2 standard deviations). Boxplots show the median, first and third quartiles, with whiskers showing the 5th and 95th percentiles.
Extended Data Fig. 4 Clustering of the morphological profiles.
(a) Heatmap of morphological profiles. Each row represents the morphological profile of a genotype. Ancestor (red) and compensated (blue) strains are marked along the column next to the dendrogram (labeled as column an-ev). The column left to the heatmap (labeled as column WT) indicates wild-type (red) and control evolved strains (blue). Columns of the heatmap are the first eight principal components with colors representing the principal component scores. The dendrogram is the result of hierarchical clustering with red boxes representing 11 significant clusters (see Methods). Representative images of the wild-type (WT) and strains from the two clusters: i) containing cells with small bud angle (esc2-ev3, Cluster #11) and ii) cells with enlarged bud size relative to mother cell size (mms22-ev1, Cluster #02), are shown. We note that the strains harboring deletion of DNA damage responding genes are 16.5-fold enriched in the latter cluster (GO:0006974, Fisher’s exact test, P < 2 ×10−16, Supplementary Data 3). Cell wall and nuclei are colored with green and red, respectively. Scale bar (on image of WT) represents 5 μm distance. (b) Dendrogram showing hierarchical clustering of genotypes based on single-cell morphology profiles. The same dendrogram as in panel (a), but also showing the names of the strains and the approximately unbiased probability values (AU p-value) for each cluster. AU p-values were used to define statistically significant clusters (Cluster #01-11) indicated by red boxes (for further details, see Methods). For further information on the clusters, see Supplementary Data 3. High-resolution image of Extended Data Fig. 4 can be found at https://figshare.com/s/a5f1571eb8cc5bada89b.
Extended Data Fig. 5 Morphological changes are specific to compensatory evolution.
(a-b-c-d) Evolved control strains show limited change in cellular morphology. Distribution of cell size (a), cell elongation (b) and neck position (c) for the evolved controls (initiated from the wild-type background, WTevo) and compensated strains (KOevo). Each dot represents average trait value for an individual strain. Changes of the above parameters in the evolved controls are negligible in comparison to a large number of compensated strains. Horizontal line and grey area denote the average value and average ± 2 standard deviation of the wild-type replicates, respectively. Morphological traits correspond to the same CalMorph parameters as in Fig. 1c. (d) Distribution of Euclidean distance (from the wild-type) of the control evolved (WTevo) and compensated strains (KOevo). Degree of morphological changes between the wild type (WT) and evolved controls is smaller than between the WT and most of the compensated strains (Brunner-Munzel test, P = 3 ×10−14). Degree of morphological change is measured by Euclidean distance between morphological profiles (see Methods). Red dots and error bars show the average and 95% confidence interval for the two strain sets. (e) Morphological divergence during compensatory evolution is independent of the number of accumulated mutations. The figure shows the Euclidean distance of the 18 compensated strains from their corresponding ancestors as a function of the number of mutations accumulated during the course of compensatory evolution8. The left and right panel shows the number of mutations including and excluding the synonymous ones, respectively. We found a lack of significant correlation between the number of accumulated mutations and the overall morphological distance, indicating that large morphological changes are often accessible in a few mutational steps.
Extended Data Fig. 6 Larger field of views for microscopy images.
(a) Similar cellular morphology of compensated strains and natural isolates. The figure shows wider field of views for Fig. 2c. Images show pairs of compensated and natural strains that display similar morphological trait combinations (cell wall and nuclei are colored with green and red, respectively): (i) large cells with normal, wild-type-like elongation: xrs2-ev1, OS_1586 isolate from tree leaves, (ii) large round cells: vid22-ev2, OS_755 wine yeast isolate, (iii) small round cells: med1-ev4, OS_675 isolate from human blood. Scale bar represents 10 μm. (b) Synergistic epistasis underlying morphological changes in a compensated strain of Δrpb9. The figure shows wider field of view images for Fig. 5a. Images show 5 selected genotypes, including the wild-type (WT), two single mutants (Δrpb9 and Δwhi2) and a reconstructed double mutant (Δrpb9 + Δwhi2). The fifth genotype is the compensated strain of Δrpb9 (rpb9-ev2) that harbors the whi2S133* loss-of-function allele. Cell wall and nuclei are colored with green and blue, respectively.
Extended Data Fig. 7 Cell morphology progression through the cell cycle.
(a) Pearson’s correlation between cell elongation and G2 percentage (as measured by flow-cytometry). Cell elongation corresponds to CalMorph trait C115-A. WT denotes the wild-type strain. Ancestors and compensated strains are colored by red and blue, respectively. Dashed line represents the average of the WT. Grey area represents the WT average ± 2 standard deviations. We estimated standard deviation using the pool of strainwise centered replicate measurements of all investigated strains. (b) Scheme of bud growth stages through the cell cycle. (c) Plot shows cell size in different cell cycle stages of the 10 largest compensated strains. Importantly, genotypes with large mother cells also have larger buds than that of the wild–type (red line). Note that the extent of cell size increase throughout the cell cycle stages varies somewhat across the compensated strains. (d) Compensated strains displaying the most elongated mother cells reach their elongated shape during the G2/M and cytokinesis phase of the bud growth. Note that several strains show more intense bud elongation than the wild type. Size of the mother cell and bud corresponds to CalMorph traits C11-1 and C11-2, respectively. Elongation of the mother cell and bud corresponds to CalMorph traits C114 and C115, respectively. Cell cycle stages G1, G2/M and cytokinesis indicated on the plots correspond to stages A, A1B and C of the CalMorph software, respectively.
Extended Data Fig. 8 Multicellular morphologies of compensated strains.
(a-b-c) Systematic screening of multicellular morphology. Barplots show the relative invasiveness (a), the relative settling score (b) and the relative biofilm area (c) of the compensated strains (initiated from knockout backgrounds, left panel) and control evolved strains (initiated from WT, right panel), respectively. Relative invasiveness score was calculated by normalizing the invasiveness score of the strains to that of the positive control strain (sigma1278b). Relative settling score (a proxy of cell aggregation) was calculated by normalizing the settling of the strains to that of the wild type strain. Relative biofilm area was calculated by normalizing the biofilm area of the strains to that of the WT. Orange color marks those compensated strains that display the corresponding trait (see Methods). (d) Imaging multicellular aggregation. The label-free microscopy images shows wider field of views for Fig. 3e, involving clump-forming compensated strains and the non-clumping WT. (e) Flocculation assay. Heatmap on the left summarizes the response of multicellular clumps to a deflocculation agent (4 mM EDTA) that can disrupt clumps formed via Ca2+-dependent flocculation (see Methods). Deflocculation resulted in clear separation of the multicellular flocs into single / few cells (green) in a well-flocculating positive control strain (OS_1189 soil isolate, described in a previous study28). In contrast, there was no obvious change in the phenotypes of the compensated strains forming multicellular aggregates (red). Compensated strains were grouped into 3 different classes: +++/++/+ show the largest/medium-sized/smallest multicellular clumps, respectively. Microscopic images on the right show the deflocculation assay of two representative compensated strains that displayed significant settling (bub3-ev2 and rpb9-ev3), along with a flocculation positive strain (OS_1189). For microscopy analysis of the flocculation positive control strain and the compensated strains, a 10x and a 20x objective was used, respectively. Scale bar represents 50 μm distance.
Extended Data Fig. 9 Analyzing invasive growth phenotype of bub3-ev3 and natural yeast isolates.
(a) Invasive growth assay of 29 haploid natural yeast isolates. Natural baker’s yeast isolates were selected from a previous study28 and represent several phylogenetic clades (N = 8) and ecological origins (N = 10), indicated on the left panel. Relative invasiveness score (right panel) was calculated by normalizing the invasiveness of the strains to the mean of the positive control strain (sigma 1278b). The black cross and the point-range represent the mean and the standard error of the invasiveness score of at least four biological replicates (separate grey points). The red dashed line mark the mean invasiveness score of the compensated strain (bub3-ev3) that displays the strongest invasive growth phenotype. For further details, see Methods. For strain abbreviations, see Supplementary Data 5. (b) Measuring the activity of the filamentous growth pathway. Boxplot shows the activity of the FRE-lacZ reporter across several genotypes including WT, bub3-ev3 line, and a positive control strain (sigma 1278b). Activity of the FRE (Tec1p-dependent filamentous response element) gives information about activity of the filamentous growth pathway. The level of the filamentous response was estimated by measuring the β-galactosidase activity on protein extracts of yeast colonies after 3 days of incubation. To assess β-galactosidase activity, an established ONPG assay was used. Relative FRE-lacZ activity was calculated by normalizing the Miller Units of the investigated genotypes to that of the WT. Boxplots show the median, first and third quartiles, with whiskers showing the 5th and 95th percentiles of at least four biological replicates for each of the genotypes. Significant differences were assessed by two-sided Student’s t-tests (***/**** indicates P < 0.001/0.0001). The P values are 5.3 ×10−4 and 4.2 ×10−6 for comparing WT with bub3-ev3 and sigma 1278b, respectively.
Extended Data Fig. 10 Mutation in SWE1 partially compensates the fitness defect of the Δbub3 ancestor strain.
Boxplot shows the relative fitness across several genotypes, including wild-type (WT), the ancestor (bub3-an) and a compensated strain of Δbub3 (bub3-ev3), and strains harboring the reconstructed SWE1Y332S mutant allele. As a proxy for fitness, colony size after 72 h of incubation on solid medium was measured as previously8. Briefly, ordered arrays of strains at 768-density were spotted onto YPD solid medium with medium-density (2%) agar. After 48 h of acclimatization to the medium at 30 °C, plates were replicated again onto the same medium. Digital images of the plates were taken with a camera after 72 h of incubation at 30 °C. The images were then processed to calculate colony sizes, after correcting for potential systematic biases8. Genotype fitness was estimated by the mean colony size of six biological replicates (i.e. six independent colonies). Relative fitness was calculated by normalizing the absolute colony sizes (see Methods) to that of the wild type strain. Significant differences were assessed by two-sided Wilcoxon rank-sum tests (**** indicates P < 0.0001, ns = non-significant). The P values are 0.15 and 3.11 ×10−28 for comparing WT with WT + SWE1Y332S and bub3-an, respectively, while the P values are 6.92 ×10−13 and 4.16 ×10−8 for comparing bub3-an with bub3-ev3 and bub3-an + SWE1Y332S, respectively. Boxplots show the median, first and third quartiles, with whiskers showing the 5th and 95th percentiles.
Supplementary information
Supplementary Information
Supplementary Protocols, Data 8 and References.
Supplementary Table 1
Supplementary data used in this study.
Rights and permissions
About this article
Cite this article
Farkas, Z., Kovács, K., Sarkadi, Z. et al. Gene loss and compensatory evolution promotes the emergence of morphological novelties in budding yeast. Nat Ecol Evol 6, 763–773 (2022). https://doi.org/10.1038/s41559-022-01730-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-022-01730-1
This article is cited by
-
Deep Conservation and Unexpected Evolutionary History of Neighboring lncRNAs MALAT1 and NEAT1
Journal of Molecular Evolution (2024)
-
Advances in genome sequencing reveal changes in gene content that contribute to arthropod macroevolution
Development Genes and Evolution (2023)
-
Turning gene loss into phenotypic gain
Nature Reviews Genetics (2022)