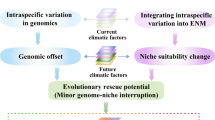
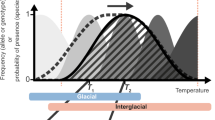
Abstract
Climate change is a threat to biodiversity. One way that this threat manifests is through pronounced shifts in the geographical range of species over time. To predict these shifts, researchers have primarily used species distribution models. However, these models are based on assumptions of niche conservatism and do not consider evolutionary processes, potentially limiting their accuracy and value. To incorporate evolution into the prediction of species’ responses to climate change, researchers have turned to landscape genomic data and examined information about local genetic adaptation using climate models. Although this is an important advancement, this approach currently does not include other evolutionary processes—such as gene flow, population dispersal and genomic load—that are critical for predicting the fate of species across the landscape. Here, we briefly review the current practices for the use of species distribution models and for incorporating local adaptation. We next discuss the rationale and theory for considering additional processes, reviewing how they can be incorporated into studies of species’ responses to climate change. We summarize with a conceptual framework of how manifold layers of information can be combined to predict the potential response of specific populations to climate change. We illustrate all of the topics using an exemplar dataset and provide the source code as potential tutorials. This Perspective is intended to be a step towards a more comprehensive integration of population genomics with climate change science.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The exemplar mexicana data used in all analyses are available at Zenodo (https://doi.org/10.5281/zenodo.4746517).
Code availability
The Markdown file is available as Supplementary Information. All R code is also available at Zenodo (https://doi.org/10.5281/zenodo.4746517).
References
Foden, W. B. et al. Climate change vulnerability assessment of species. Wiley Interdiscip. Rev. Clim. Change 10, e551 (2019).
Parmesan, C. & Yohe, G. A globally coherent fingerprint of climate change. Nature 421, 37–42 (2003).
Parmesan, C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (2006).
Walther, G.-R. et al. Ecological responses to recent climate change. Nature 416, 389–395 (2002).
Feeley, K. J., Bravo-Avila, C., Fadrique, B., Perez, T. M. & Zuleta, D. Climate-driven changes in the composition of New World plant communities. Nat. Clim. Change 10, 965–970 (2020).
Román-Palacios, C. & Wiens, J. J. Recent responses to climate change reveal the drivers of species extinction and survival. Proc. Natl Acad. Sci. USA 117, 4211–4217 (2020).
Dyderski, M. K., Paź, S., Frelich, L. E. & Jagodziński, A. M. How much does climate change threaten European forest tree species distributions? Glob. Change Biol. 24, 1150–1163 (2018).
Peterson, A. T. et al. Ecological Niches and Geographic Distributions (Princeton Univ. Press, 2011).
Thuiller, W., Guéguen, M., Renaud, J., Karger, D. N. & Zimmermann, N. E. Uncertainty in ensembles of global biodiversity scenarios. Nat. Commun. 10, 1446 (2019).
Fourcade, Y., Besnard, A. G. & Secondi, J. Evaluating interspecific niche overlaps in environmental and geographic spaces to assess the value of umbrella species. J. Avian Biol. 48, 1563–1574 (2017).
Feeley, K. J., Rehm, E. M. & Machovina, B. perspective: The responses of tropical forest species to global climate change: acclimate, adapt, migrate, or go extinct? Front. Biogeogr. 4, 69–84 (2012).
Razgour, O. et al. An integrated framework to identify wildlife populations under threat from climate change. Mol. Ecol. Resour. 18, 18–31 (2018).
Fitzpatrick, M. C. & Keller, S. R. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18, 1–16 (2015).
Exposito-Alonso, M. et al. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nat. Ecol. Evol. 2, 352–358 (2018).
Exposito-Alonso, M., Burbano, H. A., Bossdorf, O., Nielsen, R. & Weigel, D. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 573, 126–129 (2019).
Reside, A. E., Butt, N. & Adams, V. M. Adapting systematic conservation planning for climate change. Biodivers. Conserv. 27, 1–29 (2018).
Brown, J. L. et al. Predicting the genetic consequences of future climate change: the power of coupling spatial demography, the coalescent, and historical landscape changes. Am. J. Bot. 103, 153–163 (2016).
Waldvogel, A. et al. Evolutionary genomics can improve prediction of species’ responses to climate change. Evoution Lett. 4, 4–18 (2019).
Allendorf, F. W., Hohenlohe, P. A. & Luikart, G. Genomics and the future of conservation genetics. Nat. Rev. Genet. 11, 697–709 (2010).
Barbosa, S. et al. Integrative approaches to guide conservation decisions: using genomics to define conservation units and functional corridors. Mol. Ecol. 27, 3452–3465 (2018).
Nadeau, C. P. & Urban, M. C. Eco-evolution on the edge during climate change. Ecography 42, 1280–1297 (2019).
Hällfors, M. H. et al. Addressing potential local adaptation in species distribution models: Implications for conservation under climate change. Ecol. Appl. 26, 1154–1169 (2016).
Capblancq, T., Fitzpatrick, M. C., Bay, R. A., Exposito-Alonso, M. & Keller, S. R. Genomic prediction of (mal)adaptation across current and future climatic landscapes. Annu. Rev. Ecol. Evol. Syst. 51, 245–269 (2020).
Rellstab, C., Dauphin, B. & Exposito-Alonso, M. Prospects and limitations of genomic offset in conservation management. Evol. Appl. 14, 1202–1212 (2021).
Soberón, J. M. Niche and area of distribution modeling: a population ecology perspective. Ecography 33, 159–167 (2010).
Araújo, M. B. & Peterson, A. T. Uses and misuses of bioclimatic envelope modeling. Ecology 93, 1527–1539 (2012).
Wiens, J. A., Stralberg, D., Jongsomjit, D., Howell, C. A. & Snyder, M. A. Niches, models, and climate change: assessing the assumptions and uncertainties. Proc. Natl Acad. Sci. USA 106, 19729–19736 (2009).
Collart, F., Hedenäs, L., Broennimann, O., Guisan, A. & Vanderpoorten, A. Intraspecific differentiation: Implications for niche and distribution modelling. J. Biogeogr. 48, 415–426 (2020).
Benito Garzón, M., Robson, T. M. & Hampe, A. ΔTraitSDMs: species distribution models that account for local adaptation and phenotypic plasticity. N. Phytol. 222, 1757–1765 (2019).
Frichot, E., Schoville, S. D., Bouchard, G. & François, O. Testing for association between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 30, 1687–1699 (2013).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 12, 246 (2011).
Ikeda, D. H. et al. Genetically informed ecological niche models improve climate change predictions. Glob. Change Biol. 23, 164–176 (2017).
Jay, F. et al. Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 21, 2354–2368 (2012).
Razgour, O. et al. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl Acad. Sci. USA 116, 10418–10423 (2019).
Gotelli, N. J. & Stanton-Geddes, J. Climate change, genetic markers and species distribution modelling. J. Biogeogr. 42, 1577–1585 (2015).
Pyhäjärvi, T., Hufford, M. B., Mezmouk, S. & Ross-Ibarra, J. Complex patterns of local adaptation in teosinte. Genome Biol. Evol. 5, 1594–1609 (2013).
Aguirre-Liguori, J. A. et al. Connecting genomic patterns of local adaptation and niche suitability in teosintes. Mol. Ecol. 26, 4226–4240 (2017).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Kawecki, T. J. & Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241 (2004).
Leimu, R. & Fischer, M. A meta-analysis of local adaptation in plants. PLoS ONE 3, e4010 (2008).
de Villemereuil, P. & Gaggiotti, O. E. A new FST-based method to uncover local adaptation using environmental variables. Methods Ecol. Evol. 6, 1248–1258 (2015).
Coop, G. M., Witonsky, D., Di Rienzo, A. & Pritchard, J. K. Using environmental correlations to identify loci underlying local adaptation. Genetics 185, 1411–1423 (2010).
Gautier, M. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 201, 1555–1579 (2015).
De Mita, S. et al. Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol. Ecol. 22, 1383–1399 (2013).
Schoville, S. D. et al. Adaptive genetic variation on the landscape: methods and Cases. Annu. Rev. Ecol. Evol. Syst. 43, 23–43 (2012).
Tiffin, P. & Ross-Ibarra, J. Advances and limits of using population genetics to understand local adaptation. Trends Ecol. Evol. 29, 673–680 (2014).
Forester, B. R., Lasky, J. R., Wagner, H. H. & Urban, D. L. Comparing methods for detecting multilocus adaptation with multivariate genotype–environment associations. Mol. Ecol. 27, 2215–2233 (2018).
Fitzpatrick, M. C., Chhatre, V. E., Soolanayakanahally, R. Y. & Keller, S. R. Experimental support for genomic prediction of climate maladaptation using the machine learning approach Gradient Forests. Mol. Ecol. Resour. https://doi.org/10.1111/1755-0998.13374 (2021).
Gougherty, A. V., Keller, S. R. & Fitzpatrick, M. C. Maladaptation, migration and extirpation fuel climate change risk in a forest tree species. Nat. Clim. Change 11, 166–171 (2021).
Fitzpatrick, M. C., Keller, S. R. & Lotterhos, K. E. Comment on ‘Genomic signals of selection predict climate-driven population declines in a migratory bird’. Science 361, eaat7279 (2018).
Booker, T. R., Yeaman, S. & Whitlock, M. C. Variation in recombination rate affects detection of outliers in genome scans under neutrality. Mol. Ecol. 29, 4274–4279 (2020).
Rhoné, B. et al. Pearl millet genomic vulnerability to climate change in West Africa highlights the need for regional collaboration. Nat. Commun. 11, 5274 (2020).
Bascompte, J., García, M. B., Ortega, R., Rezende, E. L. & Pironon, S. Mutualistic interactions reshuffle the effects of climate change on plants across the tree of life. Sci. Adv. 5, eaav2539 (2019).
Hampe, A. & Petit, R. J. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8, 461–467 (2005).
Sexton, J. P., Strauss, S. Y. & Rice, K. J. Gene flow increases fitness at the warm edge of a species’ range. Proc. Natl Acad. Sci. USA 108, 11704–11709 (2011).
Hufford, M. B. et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9, e1003477 (2013).
Pease, J. B., Haak, D. C., Hahn, M. W. & Moyle, L. C. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016).
Figueiró, H. V. et al. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci. Adv. 3, e1700299 (2017).
Bontrager, M. & Angert, A. L. Gene flow improves fitness at a range edge under climate change. Evol. Lett. 3, 55–68 (2019).
Todesco, M. et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584, 602–607 (2020).
Le Corre, V., Siol, M., Vigouroux, Y., Tenaillon, M. I. & Délye, C. Adaptive introgression from maize has facilitated the establishment of teosinte as a noxious weed in Europe. Proc. Natl Acad. Sci. USA 117, 25618–25627 (2020).
Oziolor, E. et al. Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science 364, 455–457 (2019).
Bolnick, D. I. & Nosil, P. Natural selection in populations subject to a migration load. Evolution 61, 2229–2243 (2007).
Eckert, C. G., Samis, K. E. & Lougheed, S. C. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188 (2008).
Sexton, J. P., McInyre, P. J., Angert, A. L. & Rice, K. J. Evolution and ecology of species range limits. Annu. Rev. Ecol. Evol. Syst. 40, 415–436 (2009).
Brady, S. P. et al. Causes of maladaptation. Evol. Appl. 12, 1229–1242 (2019).
Micheletti, S. J. & Storfer, A. Mixed support for gene flow as a constraint to local adaptation and contributor to the limited geographic range of an endemic salamander. Mol. Ecol. 29, 4091–4101 (2020).
Sagarin, R. D. & Gaines, S. D. The ‘abundant centre’ distribution: to what extent is it a biogeographical rule? Ecol. Lett. 5, 137–147 (2002).
Sagarin, R. D., Gaines, S. D. & Gaylord, B. Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends Ecol. Evol. 21, 524–530 (2006).
Fedorka, K. M., Winterhalter, W. E., Shaw, K. L., Brogan, W. R. & Mousseau, T. A. The role of gene flow asymmetry along an environmental gradient in constraining local adaptation and range expansion. J. Evol. Biol. 25, 1676–1685 (2012).
Nosil, P., Harmon, L. J. & Seehausen, O. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol. 24, 145–156 (2009).
Cenzer, M. L. Adaptation to an invasive host is driving the loss of a native ecotype. Evolution 70, 2296–2307 (2016).
Hengeveld, R. & Haeck, J. The distribution of abundance. I. Measurements. J. Biogeogr. 9, 303–316 (1982).
Farkas, T. E., Mononen, T., Comeault, A. A., Hanski, I. & Nosil, P. Evolution of camouflage drives rapid ecological change in an insect community. Curr. Biol. 23, 1835–1843 (2013).
Excoffier, L. & Foll, M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27, 1332–1334 (2011).
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., Sousa, V. C. & Foll, M. Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 (2013).
Wright, S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In Proc. Sixth International Congress of Genetics Vol. 1, 356–366 (1932).
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Nei, M. & Chesser, R. K. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 47, 253–259 (1983).
Yeaman, S. & Otto, S. P. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65, 2123–2129 (2011).
Feder, J. L., Flaxman, S. M., Egan, S. P., Comeault, A. A. & Nosil, P. Geographic mode of speciation and genomic divergence. Annu. Rev. Ecol. Evol. Syst. 44, 73–97 (2013).
Endler, J. Gene Flow and population differentiation. Science 179, 243–250 (1973).
Rehm, E. M., Olivas, P., Stroud, J. & Feeley, K. J. Losing your edge: climate change and the conservation value of range-edge populations. Ecol. Evol. 5, 4315–4326 (2015).
Chen, I. C., Hill, J. K., Ohlemüller, R., Roy, D. B. & Thomas, C. D. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011).
Chen, I. C. et al. Elevation increases in moth assemblages over 42 years on a tropical mountain. Proc. Natl Acad. Sci. USA 106, 1479–1483 (2009).
McRae, B. H., Dickson, B. G., Keitt, T. H. & Shah, V. B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89, 2712–2724 (2008).
Aguirre-Liguori, J. A., Ramírez-Barahona, S., Tiffin, P. & Eguiarte, L. E. Climate change is predicted to disrupt patterns of local adaptation in wild and cultivated maize. Proc. R. Soc. B 286, 20190486 (2019).
Kling, M. M. & Ackerly, D. D. Global wind patterns shape genetic differentiation, asymmetric gene flow, and genetic diversity in trees. Proc. Natl Acad. Sci. USA 118, e2017317118 (2021).
Henn, B. M. et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl Acad. Sci. USA 113, 440–449 (2016).
Gaut, B. S., Seymour, D. K., Liu, Q. & Zhou, Y. Demography and its effects on genomic variation in crop domestication. Nat. Plants 4, 512–520 (2018).
Frankham, R. Genetics and extinction. Biol. Conserv. 126, 131–140 (2005).
Choi, Y., Sims, G., Murphy, S., Miller, J. & Chan, A. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7, e46688 (2012).
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Davydov, E. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6, e1001025 (2010).
Yang, J. et al. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 13, e1007019 (2017).
Willi, Y., Fracassetti, M., Zoller, S. & Van Buskirk, J. Accumulation of mutational load at the edges of a species range. Mol. Biol. Evol. 35, 781–791 (2018).
Koski, M. H., Layman, N. C., Prior, C. J., Busch, J. W. & Galloway, L. F. Selfing ability and drift load evolve with range expansion. Evol. Lett. 3, 500–512 (2019).
Micheletti, S. J. & Storfer, A. A test of the central-marginal hypothesis using population genetics and ecological niche modelling in an endemic salamander (Ambystoma barbouri). Mol. Ecol. 24, 967–979 (2015).
Peischl, S. & Excoffier, L. Expansion load: recessive mutations and the role of standing genetic variation. Mol. Ecol. 24, 2084–2094 (2015).
Braasch, J. & Barker, B. S. Expansion history and environmental suitability shape effective population size in a plant invasion. Mol. Ecol. 28, 2546–2558 (2019).
Perrier, A., Sánchez-Castro, D. & Willi, Y. Expressed mutational load increases toward the edge of a species’ geographic range. Evolution 74, 1711–1723 (2020).
Zhou, Y. et al. The population genetics of structural variants in grapevine domestication. Nat. Plants 5, 965–979 (2019).
Peischl, S., Kirkpatrick, M. & Excoffier, L. Expansion load and the evolutionary dynamics of a species range. Am. Nat. 185, 81–93 (2015).
Excoffier, L., Foll, M. & Petit, R. J. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40, 481–501 (2009).
Lira-Noriega, A. & Manthey, J. D. Relationship of genetic diversity and niche centrality: a survey and analysis. Evolution 68, 1082–1093 (2014).
Bay, R. A. et al. Genomic signals of selection predict climate-driven population declines in a migratory bird. Science 359, 83–86 (2018).
Ruegg, K. et al. Ecological genomics predicts climate vulnerability in an endangered southwestern songbird. Ecol. Lett. 21, 1085–1096 (2018).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Acknowledgements
The study was supported by a UC-Mexus postdoctoral fellowship to J.A.A.-L., National Science Foundation grant no. 1741627 to B.S.G. and CONACyT Ciencia de Frontera 2019 grant no. 263962 to S.R.-B.
Author information
Authors and Affiliations
Contributions
J.A.A.-L., S.R.-B. and B.S.G shaped ideas and content, discussed the results and wrote the manuscript. J.A.A.-L. wrote the code, and S.R.-B. and J.A.A.-L. constructed the Markdown file.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Ecology & Evolution thanks Matthew Fitzpatrick, Ann-Marie Waldvogel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary protocols, containing the description and code used to perform all of the analyses in the manuscript, and Supplementary Figs. 1–15.
Rights and permissions
About this article
Cite this article
Aguirre-Liguori, J.A., Ramírez-Barahona, S. & Gaut, B.S. The evolutionary genomics of species’ responses to climate change. Nat Ecol Evol 5, 1350–1360 (2021). https://doi.org/10.1038/s41559-021-01526-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-021-01526-9
This article is cited by
-
Genomics for monitoring and understanding species responses to global climate change
Nature Reviews Genetics (2024)
-
Genomic evidence for rediploidization and adaptive evolution following the whole-genome triplication
Nature Communications (2024)
-
Genomics empowering conservation action and improvement of celery in the face of climate change
Planta (2024)
-
Genome Scan of Rice Landrace Populations Collected Across Time Revealed Climate Changes’ Selective Footprints in the Genes Network Regulating Flowering Time
Rice (2023)
-
Key triggers of adaptive genetic variability of sessile oak [Q. petraea (Matt.) Liebl.] from the Balkan refugia: outlier detection and association of SNP loci from ddRAD-seq data
Heredity (2023)