Abstract
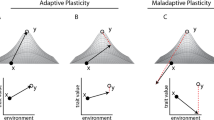
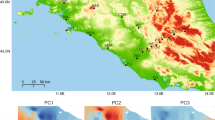
Only recently have we begun to understand the ecological and evolutionary effects of urbanization on species, with studies revealing drastic impacts on community composition, gene flow, behaviour, morphology and physiology. However, our understanding of how adaptive evolution allows species to persist, and even thrive, in urban landscapes is still nascent. Here, we examine phenotypic, genomic and regulatory impacts of urbanization on a widespread lizard, the Puerto Rican crested anole (Anolis cristatellus). We find that urban lizards endure higher environmental temperatures and display greater heat tolerance than their forest counterparts. A single non-synonymous polymorphism within a protein synthesis gene (RARS) is associated with heat tolerance plasticity within urban heat islands and displays parallel signatures of selection in cities. Additionally, we identify groups of differentially expressed genes between habitats showing elevated genetic divergence in multiple urban–forest comparisons. These genes display evidence of adaptive regulatory evolution within cities and disproportionately cluster within regulatory modules associated with heat tolerance. This study provides evidence of temperature-mediated selection in urban heat islands with repeatable impacts on physiological evolution at multiple levels of biological hierarchy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequence and metadata associated with this study have been deposited at NCBI under project no. PRJNA592594.
References
Gould, S. J. Wonderful Life: The Burgess Shale and the Nature of History (W.W. Norton & Co., 1990).
Johnson, M. T. & Munshi-South, J. Evolution of life in urban environments. Science 358, eaam8327 (2017).
Fernández-Juricic, E. Bird community composition patterns in urban parks of Madrid: the role of age, size and isolation. Ecol. Res. 15, 373–383 (2000).
Munshi-South, J. Urban landscape genetics: canopy cover predicts gene flow between white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 21, 1360–1378 (2012).
Beninde, J. et al. Cityscape genetics: structural versus functional connectivity of an urban lizard population. Mol. Ecol. 25, 4984–5000 (2016).
Miranda, A. C., Schielzeth, H., Sonntag, T. & Partecke, J. Urbanization and its effects on personality traits: a result of microevolution or phenotypic plasticity? Glob. Change Biol. 19, 2634–2644 (2013).
van Dongen, W. F. D., Robinson, R. W., Weston, M. A., Mulder, R. A. & Guay, P. Variation at the DRD4 locus is associated with wariness and local site selection in urban black swans. BMC Evol. Biol. 15, 253 (2015).
Badyaev, A. V., Young, R. L., Oh, K. P. & Addison, C. Evolution on a local scale: developmental, functional, and genetic bases of divergence in bill form and associated song structure between adjecent habitats. Evolution 62, 1951–1964 (2008).
Winchell, K. M., Reynolds, R. G., Prado-Irwin, S. R., Puente-Rolón, A. R. & Revell, L. J. Phenotypic shifts in urban areas in the tropical lizard Anolis cristatellus. Evolution 70, 1009–1022 (2016).
Brans, K. I. et al. Eco-evolutionary dynamics in urbanized landscapes: evolution, species sorting and the change in zooplankton body size along urbanization gradients. Philos. Trans. R. Soc. B 372, 20160030 (2016).
Diamond, S. E., Chick, L., Perez, A. B. E., Strickler, S. A. & Martin, R. A. Rapid evolution of ant thermal tolerance across an urban-rural temperature cline. Biol. J. Linn. Soc. 121, 248–257 (2017).
Harris, S. E. & Munshi-South, J. Signatures of positive selection and local adaptation to urbanization in white-footed mice (Peromyscus leucopus). Mol. Ecol. 26, 6336–6350 (2017).
Huey, RaymondB. & Webster, P. T. Thermal biology of Anolis lizards in a complex fauna: the christatellus group on Puerto Rico. Ecology 57, 985–994 (1976).
Huey, R. B. Behavioral thermoregulation in lizards: importance of associated costs. Science 184, 1001–1002 (1974).
Winchell, K. M., Maayan, I., Fredette, J. R. & Revell, L. J. Linking locomotor performance to morphological shifts in urban lizards. Proc. R. Soc. B 285, 20180229 (2018).
Oke, T. City size and the urban heat island. Atmos. Environ. 7, 769–779 (1973).
Angiletta, M. J. et al. Urban physiology: city ants possess high heat tolerance. PLoS ONE 2, e258 (2007).
Diamond, S. E. et al. Evolution of thermal tolerance and its fitness consequences: parallel and non-parallel responses to urban heat islands across three cities. Proc. R. Soc. B 285, 20180036 (2018).
Cowles, R. & Bogert, C. A. Preliminary study of the thermal requirements of desert reptiles. Bull. Am. Mus. Nat. Hist. 83, 265–296 (1944).
Cho, H. Y. et al. Assembly of multi-tRNA synthetase complex via heterotetrameric glutathione transferase-homology domains. J. Biol. Chem. 290, 29313–29328 (2015).
Anderson, L. L., Mao, X., Scott, B. A. & Crowder, C. M. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science 323, 630–634 (2009).
Yachdav, G. et al. PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, 337–343 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Levis, N. A. & Pfennig, D. W. Evaluating ‘plasticity-first’ evolution in nature: key criteria and empirical approaches. Trends Ecol. Evol. 31, 563–574 (2016).
Muñoz, M. M. et al. Evolutionary stasis and lability in thermal physiology in a group of tropical lizards. Proc. R. Soc. B 281, 20132433 (2014).
Homer, C. G. et al. Completion of the 2011 National Land Cover Database for the conterminous United States representing a decade of land cover change information. Photogramm. Eng. Remote Sensing 81, 345–354 (2015).
Fick, S. E. & Hijmans, R. J. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Alföldi, J. et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587–591 (2011).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Jombart, T. & Ahmed, I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A. & Cresko, W. A. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140 (2013).
Weir, B. S. Genetic Data Analysis II: Methods for Discrete Population Genetic Data (Sinauer Associates, 1996).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
We thank Z. Cheviron, N. Senner, M. Stager and J. Velotta for comments and insights that were valuable in the development of this project. We are grateful to the many people who helped in the laboratory, with animal care and in the field, in particular: K. Aviles-Rodriguez, E. Boates, D. Briggs, Q. Quach and K. Schliep.
Author information
Authors and Affiliations
Contributions
S.C.C.-S. and K.M.W. conceived the project design. S.C.C.-S., K.M.W., J.F. and I.M. performed thermal experiments. K.M.W. produced common garden data. S.C.C. and R.M.S. performed RNA-seq expression analyses. S.C.C.-S. and N.C.R. performed analyses of sequence data. S.C.C.-S., K.M.W., N.C.R., J.F., I.M., R.M.S. and J.C. participated in writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplemental methods, results, discussion, Figs. 1–20 and Tables 1–4.
Rights and permissions
About this article
Cite this article
Campbell-Staton, S.C., Winchell, K.M., Rochette, N.C. et al. Parallel selection on thermal physiology facilitates repeated adaptation of city lizards to urban heat islands. Nat Ecol Evol 4, 652–658 (2020). https://doi.org/10.1038/s41559-020-1131-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-020-1131-8
This article is cited by
-
Tadpoles Develop Elevated Heat Tolerance in Urban Heat Islands Regardless of Sex
Evolutionary Biology (2024)
-
Commonly collected thermal performance data can inform species distributions in a data-limited invader
Scientific Reports (2023)
-
Urban-rural gradients: how landscape changes drive adaptive evolution of plant competitive traits
Evolutionary Ecology (2023)
-
Draft genome of six Cuban Anolis lizards and insights into genetic changes during their diversification
BMC Ecology and Evolution (2022)
-
Parallel evolution of urban–rural clines in melanism in a widespread mammal
Scientific Reports (2022)