Abstract
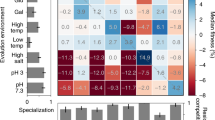
Adaptive divergence is the key evolutionary process generating biodiversity by means of natural selection. Yet, the conditions under which it can arise in the presence of gene flow remain contentious. To address this question, we subjected 132 sexually reproducing fission yeast populations, sourced from two independent genetic backgrounds, to disruptive ecological selection and manipulated the level of migration between environments. Contrary to theoretical expectations, adaptive divergence was most pronounced when migration was either absent (allopatry) or maximal (sympatry), but was much reduced at intermediate rates (parapatry and local mating). This effect was apparent across central life-history components (survival, asexual growth and mating) but differed in magnitude between ancestral genetic backgrounds. The evolution of some fitness components was constrained by pervasive negative correlations (trade-off between asexual growth and mating), while others changed direction under the influence of migration (for example, survival and mating). In allopatry, adaptive divergence was mainly conferred by standing genetic variation and resulted in ecological specialization. In sympatry, divergence was mainly mediated by novel mutations enriched in a subset of genes and was characterized by the repeated emergence of two strategies: an ecological generalist and an asexual growth specialist. Multiple loci showed consistent evidence for antagonistic pleiotropy across migration treatments providing a conceptual link between adaptation and divergence. This evolve-and-resequence experiment shows that rapid ecological differentiation can arise even under high rates of gene flow. It further highlights that adaptive trajectories are governed by complex interactions of gene flow, ancestral variation and genetic correlations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All of the data generated for this study are archived in the Sequence Read Archive (under BioProject ID PRJNA604890) at the National Centre for Biotechnology Information (www.ncbi.nlm.nih.gov/sra).
Code availability
All of the code used for the analyses, fitness data and a list of genetic variants (in .vcf format) are available at https://github.com/EvoBioWolf/SchPom_Exp_AdaptDiv and Zenodo (https://doi.org/10.5281/zenodo.4133489)89.
References
Darwin, C. & Wallace, A. R. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. Zool. J. Linn. Soc. 3, 45–62 (1858).
Schluter, D. Evidence for ecological speciation and its alternative. Science 323, 737–741 (2009).
Dobzhansky, T. Genetics and the Origin of Species Vol. 11 (Columbia Univ. Press, 1937).
Mayr, E. Animal Species and Evolution (Harvard Univ. Press, 1963).
Coyne, J. A. & Orr, H. A. Speciation (Sinauer, 2004).
Dettman, J. R., Sirjusingh, C., Kohn, L. M. & Anderson, J. B. Incipient speciation by divergent adaptation and antagonistic epistasis in yeast. Nature 447, 585–588 (2007).
Haldane, J. B. S. A mathematical theory of natural and artificial selection. (Part VI, Isolation.). Math. Proc. Camb. Phil. Soc. 26, 220–230 (1930).
Räsänen, K. & Hendry, A. P. Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecol. Lett. 11, 624–636 (2008).
Smadja, C. M. & Butlin, R. K. A framework for comparing processes of speciation in the presence of gene flow. Mol. Ecol. 20, 5123–5140 (2011).
Ronce, O. & Kirkpatrick, M. When sources become sinks: migrational meltdown in heterogeneous habitats. Evolution 55, 1520–1531 (2001).
Spichtig, M. & Kawecki, T. J. The maintenance (or not) of polygenic variation by soft selection in heterogeneous environments. Am. Nat. 164, 70–84 (2004).
Guillaume, F. & Whitlock, M. C. Effects of migration on the genetic covariance matrix. Evolution 61, 2398–2409 (2007).
Arnold, S. J., Bürger, R., Hohenlohe, P. A., Ajie, B. C. & Jones, A. G. Understanding the evolution and stability of the G-matrix. Evolution 62, 2451–2461 (2008).
Garant, D., Forde, S. E. & Hendry, A. P.The multifarious effects of dispersal and gene flow on contemporary adaptation. Funct. Ecol. 21, 434–443 (2006).
Nosil, P. Speciation with gene flow could be common. Mol. Ecol. 17, 2103–2106 (2008).
Dieckmann, U., Doebeli, M., Metz, J. A. J. & Tautz, D. Adaptive Speciation (Cambridge Univ. Press, 2012).
Shafer, A. B. A. & Wolf, J. B. W. Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol. Lett. 16, 940–950 (2013).
Hendry, A. P., Bolnick, D. I., Berner, D. & Peichel, C. L. Along the speciation continuum in sticklebacks. J. Fish. Biol. 75, 2000–2036 (2009).
Nosil, P. Ecological Speciation (Oxford Univ. Press, 2012).
Arnegard, M. E. et al. Genetics of ecological divergence during speciation. Nature 511, 307–311 (2014).
Seehausen, O. et al. Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192 (2014).
Wolf, J. B. W. & Ellegren, H. Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 18, 87–100 (2017).
Gray, J. C. & Goddard, M. R. Gene-flow between niches facilitates local adaptation in sexual populations. Ecol. Lett. 15, 955–962 (2012).
Soria-Carrasco, V. et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344, 738–742 (2014).
Schluter, D.Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766–1774 (1996).
Reznick, D. The structure of guppy life histories: the tradeoff between growth and reproduction. Ecology 64, 862–873 (1983).
Roff, D. A. Trade-offs between growth and reproduction: an analysis of the quantitative genetic evidence. J. Evol. Biol. 13, 434–445 (2000).
Haselhorst, M. S. H., Edwards, C. E., Rubin, M. J. & Weinig, C. Genetic architecture of life history traits and environment-specific trade-offs. Mol. Ecol. 20, 4042–4058 (2011).
Silva, F. F. G., Slotte, A., Johannessen, A., Kennedy, J. & Kjesbu, O. S. Strategies for partition between body growth and reproductive investment in migratory and stationary populations of spring-spawning Atlantic herring (Clupea harengus L.). Fish. Res. 138, 71–79 (2013).
Lande, R. Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33, 402–416 (1979).
Arnold, S. J. Constraints on phenotypic evolution. Am. Nat. 140, S85–S107 (1992).
Kryazhimskiy, S., Rice, D. P., Jerison, E. R. & Desai, M. M. Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014).
Butlin, R. K. Recombination and speciation. Mol. Ecol. 14, 2621–2635 (2005).
Kassen, R. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15, 173–190 (2002).
Levene, H. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87, 331–333 (1953).
Débarre, F. & Gandon, S. Evolution in heterogeneous environments: between soft and hard selection. Am. Nat. 177, E84–E97 (2011).
Nei, M. Molecular Evolutionary Genetics (Columbia Univ. Press, 1987).
Ratcliff, W. C., Denison, R. F., Borrello, M. & Travisano, M. Experimental evolution of multicellularity. Proc. Natl Acad. Sci. USA 109, 1595–1600 (2012).
Burke, M. K., Liti, G. & Long, A. D. Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae. Mol. Biol. Evol. 31, 3228–3239 (2014).
Franssen, S. U., Kofler, R. & Schlötterer, C. Uncovering the genetic signature of quantitative trait evolution with replicated time series data. Heredity 118, 42–51 (2017).
Behe, M. J. Experimental evolution, loss-of-function mutations, and “the first rule of adaptive evolution”. Q. Rev. Biol. 85, 419–445 (2010).
Anderson, J. T., Lee, C.-R., Rushworth, C. A., Colautti, R. I. & Mitchell-Olds, T. Genetic trade-offs and conditional neutrality contribute to local adaptation: genetic basis of local adaptation. Mol. Ecol. 22, 699–708 (2013).
Maclean, R. C. Adaptive radiation in microbial microcosms: microbial diversification. J. Evol. Biol. 18, 1376–1386 (2005).
Samani, P. & Bell, G. Experimental evolution of the grain of metabolic specialization in yeast. Ecol. Evol. 6, 3912–3922 (2016).
Savolainen, O., Lascoux, M. & Merilä, J. Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820 (2013).
Barton, N. H. & Cara, M. A. R. D. The evolution of strong reproductive isolation. Evolution 63, 1171–1190 (2009).
Flaxman, S. M., Wacholder, A. C., Feder, J. L. & Nosil, P. Theoretical models of the influence of genomic architecture on the dynamics of speciation. Mol. Ecol. 23, 4074–4088 (2014).
Lowry, D. B., Rockwood, R. C. & Willis, J. H. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution 62, 2196–2214 (2008).
Barton, N. & Bengtsson, B. O.The barrier to genetic exchange between hybridising populations. Heredity 57, 357–376 (1986).
Nicolaus, M. & Edelaar, P. Comparing the consequences of natural selection, adaptive phenotypic plasticity, and matching habitat choice for phenotype–environment matching, population genetic structure, and reproductive isolation in meta-populations. Ecol. Evol. 8, 3815–3827 (2018).
Smith, J. M. Sympatric speciation. Am. Nat. 100, 637–650 (1966).
Filchak, K. E., Roethele, J. B. & Feder, J. L. Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature 407, 739–742 (2000).
Flaxman, S. M., Feder, J. L. & Nosil, P. Genetic hitchhiking and the dynamic buildup of genomic divergence during speciation with gene flow. Evolution 67, 2577–2591 (2013).
Sexton, J. P., Hangartner, S. B. & Hoffmann, A. A. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68, 1–15 (2014).
Powell, T. H. Q. et al. Genetic divergence along the speciation continuum: the transition from host race to species in Rhagoletis (diptera: Tephritidae). Evolution 67, 2561–2576 (2013).
Roux, C. et al. Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14, e2000234 (2016).
Wright, S. Evolution in Mendelian populations. Genetics 16, 97–159 (1931).
Mayr, E. in Evolution as a Process (eds Huxley, J. S. et al.) 157–180 (Allen & Unwin, 1954).
Bulmer, M. G. Multiple niche polymorphism. Am. Nat. 106, 254–257 (1972).
Fry, J. D. Multilocus models of sympatric speciation: Bush versus Rice versus Felsenstein. Evolution 57, 1735–1746 (2003).
Nieuwenhuis, B. P. S. et al. Repeated evolution of self-compatibility for reproductive assurance. Nat. Commun. 9, 1639 (2018).
Curtsinger, J. W., Service, P. M. & Prout, T. Antagonistic pleiotropy, reversal of dominance, and genetic polymorphism. Am. Nat. 144, 210–228 (1994).
Charlesworth, B. & Hughes, K. A. in Evolutionary Genetics: from Molecules to Morphology Vol. 1 (eds Singh, R. S. & Krimbas, C. B.) 369–391 (Cambridge Univ. Press, 2000).
Phillips, P. C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 (2008).
Carter, A. J. & Nguyen, A. Q. Antagonistic pleiotropy as a widespread mechanism for the maintenance of polymorphic disease alleles. BMC Med. Genet. 12, 160 (2011).
Hedrick, P. W., Ginevan, M. E. & Ewing, E. P. Genetic polymorphism in heterogeneous environments. Annu. Rev. Ecol. Syst. 7, 1–32 (1976).
Macnair, M. R. Why the evolution of resistance to anthropogenic toxins normally involves major gene changes: the limits to natural selection. Genetica 84, 213–219 (1991).
Ono, J., Gerstein, A. C. & Otto, S. P. Widespread genetic incompatibilities between first-step mutations during parallel adaptation of Saccharomyces cerevisiae to a common environment. PLoS Biol. 15, e1002591 (2017).
Blount, Z. D., Lenski, R. E. & Losos, J. B. Contingency and determinism in evolution: replaying life’s tape. Science 362, eaam5979 (2018).
Jeffares, D. C. The natural diversity and ecology of fission yeast. Yeast 35, 253–260 (2018).
Heim, L. Construction of an h+S strain of Schizosaccharomyces pombe. Curr. Genet. 17, 13–19 (1990).
Forsburg, S. L. Schizosaccharomyces pombe strain maintenance and media. Curr. Protoc. Mol. Biol. 64, 13.15.1–13.15.5 (2003).
Ellis, B. et al. flowCore: flowCore: Basic structures for flow cytometry data. R package version 1.52.1 (2019).
Kassambara, A. & Mundt, F. factoextra: Extract and visualize the results of multivariate data analyses. R package version 3.4.4 https://rdrr.io/cran/factoextra/ (2017).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880 (2002).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Li, H. Toward better understanding of artefacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Lock, A. et al. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 47, D821–D827 (2019).
Nei, M. & Li, W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269–5273 (1979).
Weir, B. S. & Cockerham, C. C.Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Haller, B. C. & Messer, P. W. SLIM 3: forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36, 632–637 (2019).
Farlow, A. et al. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics 201, 737–744 (2015).
Munz, P., Wolf, K., Kohli, J. & Leupold, U. in Molecular Biology of the Fission Yeast 1–30 (Academic Press, 1989).
Tusso, S., Nieuwenhuis, B. P. S., Weissensteiner, B., Immler, S. & Wolf, J. B. W. Data from: Experimental evolution of adaptive divergence under varying degrees of gene flow. (Zenodo, 2020); https://doi.org/10.5281/ZENODO.4133489
Acknowledgements
We thank S. L. Ament-Velásquez, R. Butlin, U. Knief, D. Metzler, C. Peart, R. Pereira, R. Stelkens, M. Weissensteiner and members of the Immler and Wolf laboratories for providing intellectual input on the various analyses, and comments on the manuscript. We are further indebted to T. Ahmad, G. Kumpfmüller, H. Lainer and N. Zajac for help with the laboratory work, D. Scofield for bioinformatics support, and B. Haller for support with running SLiM. We further acknowledge support for genomic data generation from the SNP&SEQ Technology Platform in the National Genomics Infrastructure, Uppsala, Sweden. Flow cytometry was performed at the Core Facility Flow Cytometry at LMU. The computational infrastructure was provided by the UPPMAX Next-Generation Sequencing Cluster and Storage (UPPNEX) project funded by the Knut and Alice Wallenberg Foundation and Swedish National Infrastructure for Computing. Funding was provided to J.B.W.W. by LMU Munich, the Science of Life Laboratories National Projects and Uppsala University.
Author information
Authors and Affiliations
Contributions
S.T., B.P.S.N., S.I. and J.B.W.W. conceived of the study idea. S.T., B.P.S.N. and B.W. performed the experiments. S.T. and B.W. performed the phenotypic measurements. All analyses were performed by S.T., with contributions to the phenotypic analyses from B.P.S.N. S.T. and J.B.W.W. wrote the manuscript with input from B.P.S.N. and S.I.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Ecology & Evolution thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Distance between relative fitness as a function of migration treatment.
a. PCA using z-score normalized fitness values per genetic background. Distribution of populations are given per treatment, with pairs of populations connected by a line. b. Boxplot of Euclidian distance between pairs of connected population pairs per treatment and genetic background. For allopatric populations (Allo*), the mean distance was calculated for a subset of 100 bootstrapped combinations of all top and bottom populations. In parapatric populations, the boxplot displays the distances between connected populations (Para) and bootstrapped combinations between independent top and bottom populations as in Allo* (Para*). Pairs of parapatric and local mating population are more similar in fitness than allopatric, randomized parapatric or sympatric populations. This is consistent with a homogenizing effect of gene flow for these treatments. Boxplots description: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers.
Extended Data Fig. 2 Principle component analyses of z-score standardized log transformed relative fitness values across fitness components.
Each point represents one population or subpopulation subjected to the top (blue) or bottom (red) selection regime. Eigenvectors are indicated by arrows for each fitness component. Arrows pointing in the same direction reflect a positive relationship between components, arrows in opposite direction indicate a negative correlation or trade-off and orthogonal arrows suggest no relationship. The relative contribution to variance in PC1 and PC2 is symbolized by length of the arrow. Proportion of variance explained by each PC is shown in parenthesis for both axes. Results are shown for the α and β genetic background. Abbreviations of migration treatments as in the main manuscript.
Extended Data Fig. 3 Standing genetic variation in ancestral populations and total genetic variation per evolved population.
a. Venn diagram displaying the number of genetic variants per genetic background in the two ancestral populations at the beginning of the experiment. The left panel includes all variants, whereas the right panel is restricted to variants with allele frequency higher than 0.2. b. Boxplot with number of genetic variants per population, differentiating between standing genetic variation and new mutations emerging during the experiment. Panels include either all variants or only variants with allele frequency higher than 0.2, as well as between genetic backgrounds (α and β genetic background). The orange point shows the number of genetic variants in ancestral populations at the beginning of the experiment. At the end of the experiment we counted in total 1,472 and 1,318 genetic variants (α and β respectively) of which 1,217 and 1,061 arose de-novo during the experiment. Most variants were present at low frequencies (1,179 and 1,073 variants with a maximum frequency of 0.2), and most were limited to single populations (950 and 809 variants). At the end of the experiment, each of the 132 evolved populations had between 71 and 183 mutations. Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 4 Genetic differentiation between populations.
PCA including all populations per genetic background. The analysis was conducted including all genetic variants (upper graphs) or using only variants present at the beginning of the experiment (standing genetic variation; lower graphs). Decomposition of genetic variation across all populations using principal component analyses shows a similar distribution of populations for the genetic backgrounds across the different levels of gene flow for the first two principal components. Using all genetic variants, the two major axes of variation explained in total 17.3 % and 20.1 % of the genetic variation for the α and β background, respectively. This increased to 27.7 and 25.9% when using only standing genetic variation, however, the distribution of populations remained similar.
Extended Data Fig. 5 Dxy between populations.
Boxplot of distribution of Dxy between pairs of populations. Each point represents a Dxy value for a single pair of populations. Dxy values were calculated using all genetic variants (left) or only standing genetic variation (right). Distribution of values were divided per genetic background (α and β). For each treatment, comparisons were done between populations with the same or opposite ecological selection regime (Top - T and Bottom – B). For parapatric populations, Dxy was additionally calculated between connected population pairs (Para_Pairs). Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 6 Fst between populations.
Boxplot of distribution of Fst between pairs of populations. Each point represents an Fst value for a single pair of populations. Fst values were calculated using all genetic variants (left) or only standing genetic variation (right). Distribution of values were divided per genetic background (α and β). For each treatment, comparisons were done between populations with the same or opposite ecological selection regime (Top - T and Bottom – B). For parapatric populations, Fst was additionally calculated between connected population pairs (Para_Pairs). Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 7 Genetic divergence between Top and Bottom ecotypes.
Boxplot of genetic divergence, measured as Da (Difference between Dxy and mean π), between the top and bottom ecotypes per population. Each point represents the comparison between top and bottom ecotypes within each population. The plot is divided by treatment (Local mating and Sympatry) and by genetic background. In the α genetic background genetic divergence was higher for Sympatric populations compared with Local Mating populations, which is consistent with phenotypic data. This divergence was not observed for the β background, however the absolute phenotypic variation was lower in this genetic background (Fig. 2a). Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 8 Summary statistics of allele frequency change between subpopulations.
Data are shown for the local mating (LM) and sympatry (SYM) treatment and are displayed for both ancestral genetic backgrounds (α and β). For each of the 11 populations per treatment the allele frequencies of segregating variants were compared between subpopulations including the following comparisons. Left panel (Top-Bottom): Comparison between the top and bottom ecotypic fractions; comparison between the population pool (prior to ecological selection) with the top (Middle panel: Pool-Top) or bottom ecotypic fraction (Right panel: Pool -Bottom). For each population the proportion of genetic variants with an allele frequency shift of > 0.2 were calculated. The resulting proportion of genetic variants changing in either direction as displayed for each population in Supplementary Fig. 4 are here summarized to show the general trend. Following the colour scheme from Supplementary Fig. 4 boxplots are coloured by the fraction showing an increase in frequency, viz. blue for the top ecotype (diff_Top), red for the bottom ecotype (diff_Bottom) or orange for the pool (diff_Pool). In the α background sympatric populations showed higher similarity between the whole pool sample and the top ecotype fraction (higher proportion of variants increased in top fraction, lower proportion of variants differing between top and the whole pool sample, and higher proportion of variants differing between bottom and whole pool sample). In local mating populations, the pattern was reversed showing higher similarity with the bottom ecotype fraction. In the β background, showed a similar trend, but the difference between treatments was lower, which is consistent with no observed divergence in the phenotypic data. Significance of the difference between groups was tested using a quasibinomial model in a nested generalised lineal model with treatment and fraction as fixed variables. Significant difference between treatments and group are shown with blue and black asterisks respectively. Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 9 Distribution of effect sizes for all genetic variants.
Boxplot of distribution of maximum allele frequencies grouping variants by the predicted functional effect level. Each point represents the maximum allele frequency observed in all populations per variant. Upper and lower panels are differentiated by genetic background (α and β populations) and include all variants (left) or only variants already present in the respective ancestral population (standing genetic variation, right). Effect levels as described in method section. P-value < 0.001 (***), < 0.01 (**), or < 0.05 (*). For further details see methods. Boxplots as in Extended Data Fig. 1.
Extended Data Fig. 10 Candidate genetic variants under disruptive selection per genetic background.
Boxplot of allele frequency distribution. Only variants with significant difference between top and bottom allopatric population following from logistic regression are shown. Genetic variants are labelled with chromosome, position and alternative allele relative to the reference genome. Red stars indicate variants with a significant difference between top and bottom populations, using a quasibinomial model accounting for over dispersion in the data. Allele frequencies for ancestral populations in each genetic variant is shown with orange points. Boxplots as in Extended Data Fig. 1.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19 and Tables 1–3.
Rights and permissions
About this article
Cite this article
Tusso, S., Nieuwenhuis, B.P.S., Weissensteiner, B. et al. Experimental evolution of adaptive divergence under varying degrees of gene flow. Nat Ecol Evol 5, 338–349 (2021). https://doi.org/10.1038/s41559-020-01363-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-020-01363-2