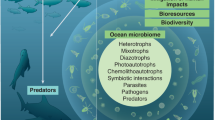
Abstract
Research into the microbiomes of natural environments is changing the way ecologists and evolutionary biologists view the importance of microorganisms in ecosystem function. This is particularly relevant in ocean environments, where microorganisms constitute the majority of biomass and control most of the major biogeochemical cycles, including those that regulate Earth’s climate. Coastal marine environments provide goods and services that are imperative to human survival and well-being (for example, fisheries and water purification), and emerging evidence indicates that these ecosystem services often depend on complex relationships between communities of microorganisms (the ‘microbiome’) and the environment or their hosts — termed the ‘holobiont’. Understanding of coastal ecosystem function must therefore be framed under the holobiont concept, whereby macroorganisms and their associated microbiomes are considered as a synergistic ecological unit. Here, we evaluate the current state of knowledge on coastal marine microbiome research and identify key questions within this growing research area. Although the list of questions is broad and ambitious, progress in the field is increasing exponentially, and the emergence of large, international collaborative networks and well-executed manipulative experiments are rapidly advancing the field of coastal marine microbiome research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Coral reef, Alexander J. Fordyce; mangrove, Michael Bradley.


Similar content being viewed by others
Data availability
The original questions for the horizon scan are available in the Supplementary Information.
Code availability
The code used to extract literature from databases is available in the Supplementary Information.
References
Liquete, C. et al. Current status and future prospects for the assessment of marine and coastal ecosystem services: a systematic review. PLoS ONE 8, e67737 (2013).
Harvell, C. et al. Emerging marine diseases — climate links and anthropogenic factors. Science 285, 1505–1510 (1999).
Penesyan, A., Kjelleberg, S. & Egan, S. Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 8, 438–459 (2010).
Dash, H. R., Mangwani, N., Chakraborty, J., Kumari, S. & Das, S. Marine bacteria: potential candidates for enhanced bioremediation. Appl. Microbiol. Biotechnol. 97, 561–571 (2013).
Hentschel, U., Piel, J., Degnan, S. M. & Taylor, M. W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641 (2012).
Seymour, J., Laverock, B., Nielsen, D., Trevathan-Tackett, S. & Macreadie, P. in Seagrasses of Australia (eds Larkum, A. W. D., Kendrick, G. A. & Ralph, P. J.) 343–392 (Springer, 2018).
Bourne, D. G., Morrow, K. M. & Webster, N. S. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340 (2016).
Hernandez-Agreda, A., Gates, R. D. & Ainsworth, T. D. Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 25, 125–140 (2016).
Marzinelli, E. M. et al. Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17, 4078–4088 (2015).
Fahimipour, A. K. et al. Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83, e03391–03316 (2017).
Tremaroli, V. & Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249 (2012).
Fernandes, N., Steinberg, P., Rusch, D., Kjelleberg, S. & Thomas, T. Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed Delisea pulchra. PLoS ONE 7, e50854 (2012).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Tout, J. et al. Variability in microbial community composition and function between different niches within a coral reef. Microb. Ecol. 67, 540–552 (2014).
Moitinho-Silva, L. et al. Integrated metabolism in sponge–microbe symbiosis revealed by genome-centered metatranscriptomics. ISME J. 11, 1651–1666 (2017).
Thompson, L. R. et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Sieber, C. M. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Ainsworth, T. D. et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274 (2015).
Spoerner, M., Wichard, T., Bachhuber, T., Stratmann, J. & Oertel, W. Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factors. J. Phycol. 48, 1433–1447 (2012).
Lee, K. S. et al. An automated Raman-based platform for the sorting of live cells by functional properties. Nat. Microbiol. 4, 1035–1048 (2019).
Lagier, J.-C. et al. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550 (2018).
Garcias-Bonet, N., Arrieta, J. M., Duarte, C. M. & Marbà, N. Nitrogen-fixing bacteria in Mediterranean seagrass (Posidonia oceanica) roots. Aquat. Bot. 131, 57–60 (2016).
Welsh, R. M. et al. Bacterial predation in a marine host-associated microbiome. ISME J. 10, 1540–1544 (2016).
Certner, R. H. & Vollmer, S. V. Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ. Microbiol. 20, 645–657 (2018).
Mejia, A. Y. et al. Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila Stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecol. Indic. 60, 1150–1163 (2016).
Glasl, B. et al. Microbiome variation in corals with distinct depth distribution ranges across a shallow–mesophotic gradient (15–85 m). Coral Reefs 36, 447–452 (2017).
Lachnit, T., Meske, D., Wahl, M., Harder, T. & Schmitz, R. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665 (2011).
Kimes, N. E. et al. The Montastraea faveolata microbiome: ecological and temporal influences on a Caribbean reef-building coral in decline. Environ. Microbiol. 15, 2082–2094 (2013).
Simister, R., Taylor, M. W., Rogers, K. M., Schupp, P. J. & Deines, P. Temporal molecular and isotopic analysis of active bacterial communities in two New Zealand sponges. FEMS Microbiol. Ecol. 85, 195–205 (2013).
Lema, K. A., Bourne, D. G. & Willis, B. L. Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695 (2014).
Gantt, S. E., López-Legentil, S. & Erwin, P. M. Stable microbial communities in the sponge Crambe crambe from inside and outside a polluted Mediterranean harbor. FEMS Microbiol. Lett. 364, fnx105 (2017).
Cúcio, C., Engelen, A. H., Costa, R. & Muyzer, G. Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7, 440 (2016).
van de Water, J. A. J. M. et al. Seasonal stability in the microbiomes of temperate gorgonians and the red coral Corallium rubrum across the Mediterranean Sea. Microb. Ecol. 75, 274–288 (2018).
Douglas, A. E. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb. Perspect. Biol. 6, a016113 (2014).
Kelly, L. W. et al. Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc. Natl Acad. Sci. USA 111, 10227–10232 (2014).
Janzen, D. H. When is it coevolution? Evolution 34, 611–612 (1980).
Thomas, T. et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 11870 (2016).
Easson, C. G. & Thacker, R. W. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5, 532 (2014).
Borges, R. M. Co-niche construction between hosts and symbionts: ideas and evidence. J. Genet. 96, 483–489 (2017).
Koskella, B., Hall, L. J. & Metcalf, C. J. E. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1, 1606 (2017).
Brooks, A. W., Kohl, K. D., Brucker, R. M., van Opstal, E. J. & Bordenstein, S. R. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 (2016).
Pollock, F. J. et al. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 4921 (2018).
Ramsby, B. D., Hoogenboom, M. O., Whalan, S. & Webster, N. S. Elevated seawater temperature disrupts the microbiome of an ecologically important bioeroding sponge. Mol. Ecol. 27, 2124–2137 (2018).
Brothers, C. J. et al. Ocean warming alters predicted microbiome functionality in a common sea urchin. Proc. R. Soc. B 285, 20180340 (2018).
Glasl, B., Smith, C. E., Bourne, D. G. & Webster, N. S. Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ 7, e6377 (2019).
Thompson, J. N. The Geographic Mosaic of Coevolution (Univ. Chicago Press, 2005).
Singh, R. P. & Reddy, C. Unraveling the functions of the macroalgal microbiome. Front. Microbiol. 6, 1488 (2016).
Garren, M. et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999 (2014).
Kubanek, J. et al. Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc. Natl Acad. Sci. USA 100, 6916–6921 (2003).
Burke, C., Steinberg, P., Rusch, D., Kjelleberg, S. & Thomas, T. Bacterial community assembly based on functional genes rather than species. Proc. Natl Acad. Sci. USA 108, 14288–14293 (2011).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012).
Rosado, P. M. et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936 (2018).
Sodré, V. et al. Physiological aspects of mangrove (Laguncularia racemosa) grown in microcosms with oil-degrading bacteria and oil contaminated sediment. Environ. Pollut. 172, 243–249 (2013).
Egan, S. et al. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. s. 37, 462–476 (2013).
Webster, N. S. & Thomas, T. The sponge hologenome. mBio 7, e00135–00116 (2016).
Meyle, J. & Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontol. 2000 69, 7–17 (2015).
Parekh, P. J., Balart, L. A. & Johnson, D. A. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin. Transl. Gastroenterol. 6, e91 (2015).
Egan, S. & Gardiner, M. Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7, 991 (2016).
Mera, H. & Bourne, D. G. Disentangling causation: complex roles of coral-associated microorganisms in disease. Environ. Microbiol. 20, 431–449 (2018).
Campbell, A. H., Harder, T., Nielsen, S., Kjelleberg, S. & Steinberg, P. D. Climate change and disease: bleaching in a chemically-defended seaweed. Glob. Change Biol. 17, 2958–2970 (2011).
Peixoto, R. S., Rosado, P. M., Leite, D. C., Rosado, A. S. & Bourne, D. G. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8, 341 (2017).
Sheaves, M. Consequences of ecological connectivity: the coastal ecosystem mosaic. Mar. Ecol. Prog. Ser. 391, 107–115 (2009).
Bates, A. E. et al. Biologists ignore ocean weather at their peril. Nature 560, 299–301 (2018).
Simon, J.-C., Marchesi, J. R., Mougel, C. & Selosse, M.-A. Host-microbiota interactions: from holobiont theory to analysis. Microbiome 7, 5 (2019).
Cuellar-Gempeler, C. & Leibold, M. A. Key colonist pools and habitat filters mediate the composition of fiddler crab-associated bacterial communities. Ecology 100, e02628 (2019).
Booth, J. M., Fusi, M., Marasco, R., Mbobo, T. & Daffonchio, D. Fiddler crab bioturbation determines consistent changes in bacterial communities across contrasting environmental conditions. Sci. Rep. 9, 3749 (2019).
Qiu, Z. et al. Future climate change is predicted to affect the microbiome and condition of habitat-forming kelp. Proc. R. Soc. B 286, 20181887 (2019).
Douglass, J. G., France, K. E., Richardson, J. P. & Duffy, J. E. Seasonal and interannual change in a Chesapeake Bay eelgrass community: insights into biotic and abiotic control of community structure. Limnol. Oceanogr. 55, 1499–1520 (2010).
Casey, J. M., Connolly, S. R. & Ainsworth, T. D. Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5, srep11903 (2015).
Campbell, A. H., Marzinelli, E. M., Gelber, J. & Steinberg, P. D. Spatial variability of microbial assemblages associated with a dominant habitat-forming seaweed. Front. Microbiol. 6, 230 (2015).
Lafferty, K. D. & Holt, R. D. How should environmental stress affect the population dynamics of disease? Ecol. Lett. 6, 654–664 (2003).
Huey, R. B. & Stevenson, R. D. Integrating thermal physiology and ecology of ectotherms: a discussion of approaches. Am. Zool. 19, 357–366 (1979).
Vega Thurber, R. et al. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163 (2009).
Fan, L., Liu, M., Simister, R., Webster, N. S. & Thomas, T. Marine microbial symbiosis heats up: the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 7, 991–1002 (2013).
Lokmer, A. & Mathias Wegner, K. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 9, 670–682 (2015).
Case, R. J. et al. Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13, 529–537 (2011).
Greff, S. et al. The interaction between the proliferating macroalga Asparagopsis taxiformis and the coral Astroides calycularis induces changes in microbiome and metabolomic fingerprints. Sci. Rep. 7, 42625 (2017).
Brown, A. L., Lipp, E. K. & Osenberg, C. W. Algae dictate multiple stressor effects on coral microbiomes. Coral Reefs 38, 229–240 (2019).
van der Heide, T. et al. A three-stage symbiosis forms the foundation of seagrass ecosystems. Science 336, 1432–1434 (2012).
Roth-Schulze, A. J. et al. The effects of warming and ocean acidification on growth, photosynthesis, and bacterial communities for the marine invasive macroalga Caulerpa taxifolia. Limnol. Oceanogr. 63, 459–471 (2018).
Banerjee, S., Schlaeppi, K. & Heijden, M. G. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576 (2018).
Jaspers, C. et al. Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Zoology 133, 81–87 (2019).
Bang, C. et al. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology 127, 1–19 (2018).
Tyagi, M., da Fonseca, M. M. R. & de Carvalho, C. C. Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22, 231–241 (2011).
Nikolopoulou, M. & Kalogerakis, N. Biostimulation strategies for fresh and chronically polluted marine environments with petroleum hydrocarbons. J. Chem. Technol. Biotechnol. 84, 802–807 (2009).
Poi, G., Aburto-Medina, A., Mok, P. C., Ball, A. S. & Shahsavari, E. Large scale bioaugmentation of soil contaminated with petroleum hydrocarbons using a mixed microbial consortium. Ecol. Eng. 102, 64–71 (2017).
Yu, K., Wong, A., Yau, K., Wong, Y. & Tam, N. Natural attenuation, biostimulation and bioaugmentation on biodegradation of polycyclic aromatic hydrocarbons (PAHs) in mangrove sediments. Mar. Pollut. Bull. 51, 1071–1077 (2005).
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I. & Dangl, J. L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163 (2017).
Slattery, J., MacFabe, D. F. & Frye, R. E. The significance of the enteric microbiome on the development of childhood disease: a review of prebiotic and probiotic therapies in disorders of childhood. Clin. Med. Insights Pediatr. 10, 91–107 (2016).
Carnevali, O., Maradonna, F. & Gioacchini, G. Integrated control of fish metabolism, wellbeing and reproduction: the role of probiotic. Aquaculture 472, 144–155 (2017).
Oppen, M. J. et al. Shifting paradigms in restoration of the world’s coral reefs. Glob. Change Biol. 23, 3437–3448 (2017).
Stewart, J. R. et al. The coastal environment and human health: microbial indicators, pathogens, sentinels and rese rvoirs. Environ. Health 7, S3 (2008).
Takemura, A. F., Chien, D. M. & Polz, M. F. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5, 38 (2014).
Ralston, E. P., Kite-Powell, H. & Beet, A. An estimate of the cost of acute food and water borne health effects from marine pathogens and toxins in the United States. J. Water Health 9, 680–694 (2011).
Lamb, J. B. et al. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733 (2017).
Halpern, B. S. et al. Spatial and temporal changes in cumulative human impacts on the world’s ocean. Nat. Commun. 6, 7615 (2015).
Labbate, M., Seymour, J. R., Lauro, F. & Brown, M. V. Anthropogenic impacts on the microbial ecology and function of aquatic environments. Front. Microbiol. 7, 1044 (2016).
Glasl, B., Webster, N. S. & Bourne, D. G. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 164, 91 (2017).
Gillings, M. R. et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279 (2015).
McLellan, S. L. & Eren, A. M. Discovering new indicators of fecal pollution. Trends Microbiol. 22, 697–706 (2014).
Fisher, J. C. et al. Comparison of sewage and animal fecal microbiomes by using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl. Environ. Microbiol. 81, 7023–7033 (2015).
Fisher, J. C., Levican, A., Figueras, M. J. & McLellan, S. L. Population dynamics and ecology of Arcobacter in sewage. Front. Microbiol. 5, 525 (2014).
Knights, D. et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011).
Roguet, A., Eren, A. M., Newton, R. J. & McLellan, S. L. Fecal source identification using random forest. Microbiome 6, 185 (2018).
Moitinho-Silva, L. et al. The sponge microbiome project. GigaScience 6, 1–7 (2017).
Tripathi, A. et al. Are microbiome studies ready for hypothesis-driven research? Curr. Opin. Microbiol. 44, 61–69 (2018).
Sutherland, W. J. et al. Identification of 100 fundamental ecological questions. J. Ecol. 101, 58–67 (2013).
Cook, C. N., Inayatullah, S., Burgman, M. A., Sutherland, W. J. & Wintle, B. A. Strategic foresight: how planning for the unpredictable can improve environmental decision-making. Trends Ecol. Evol. 29, 531–541 (2014).
Hemming, V., Burgman, M. A., Hanea, A. M., McBride, M. F. & Wintle, B. C. A practical guide to structured expert elicitation using the IDEA protocol. Methods Ecol. Evol. 9, 169–180 (2018).
Martin, T. G. et al. Eliciting expert knowledge in conservation science. Conserv. Biol. 26, 29–38 (2012).
Feehery, G. R. et al. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS ONE 8, e76096 (2013).
Eren, A. M. et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119 (2013).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Edgar, R. C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375 (2018).
Pogoreutz, C. et al. Nitrogen fixation aligns with nifH abundance and expression in two coral trophic functional groups. Front. Microbiol. 8, 1187 (2017).
Cardini, U. et al. Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching. Environ. Microbiol. 18, 2620–2633 (2016).
Glasl, B., Herndl, G. J. & Frade, P. R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292 (2016).
do Carmo, F. L. et al. Bacterial structure and characterization of plant growth promoting and oil degrading bacteria from the rhizospheres of mangrove plants. J. Microbiol. 49, 535–543 (2011).
Bourlat, S. J. et al. Genomics in marine monitoring: new opportunities for assessing marine health status. Mar. Pollut. Bull. 74, 19–31 (2013).
Acknowledgements
We thank Deakin University’s School of Life and Environmental Sciences and the Centre for Integrative Ecology for funding the workshop (New Activities Scheme 2017). B.L.’s attendance at the workshop was funded by the New Zealand Ministry of Business, Innovation and Employment Smart Ideas project UOWX1602. L.M.’s attendance was supported by an Australian Research Council Discovery Project (DP160103811). A.H.C.’s attendance was funded by the Centre for Marine Bio-Innovation at UNSW Australia. A.H.E. was supported by CCMAR/ID/16/2018, within CEECINST/00114/2018 and UID/Multi/04326/2019 financed by Fundação para a Ciência e a Tecnologia (FCT). D.D. was supported by King Abdullah University of Science and Technology through the baseline research fund and the Competetive Research Grant CRG-7-3739, Microlanding. S.M.T.-T. was supported by Deakin University’s SEBE Postdoctoral Industry Fellowship, the Mary Collins Trust and the Alfred Deakin Postdoctoral Research Fellowship. We thank J. Martiny, S. Robbins, G. Tyson, C. Weihe and K. Whiteson for their input in the horizon scanning exercise. We thank L. Koop for producing the conceptual designs.
Author information
Authors and Affiliations
Contributions
S.M.T.-T. conceived the idea; S.M.T.-T., C.D.H.S. and P.I.M. developed and led the workshop; S.M.T.-T., C.D.H.S., P.I.M., M.J.H., A.H.C., B.L., V.H.-M., J.R.S., L.F.M., T.D.A. and K.L.H. attended the workshop that structured the themes of the manuscript; S.M.T.-T., C.D.H.S., P.I.M., M.J.H., A.H.C., B.L., V.H.-M., J.R.S., L.F.M., T.D.A., K.L.H., A.F., D.D., S.E., A.H.E., M.F., T.T., L.V., A.H.-A., H.M.G., E.M.M. and P.D.S. contributed to the questions and wrote the manuscript; A.F., V.H.-M., S.M.T.-T. and L.H. contributed to the literature search.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, methods and Data 1.
Rights and permissions
About this article
Cite this article
Trevathan-Tackett, S.M., Sherman, C.D., Huggett, M.J. et al. A horizon scan of priorities for coastal marine microbiome research. Nat Ecol Evol 3, 1509–1520 (2019). https://doi.org/10.1038/s41559-019-0999-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0999-7
This article is cited by
-
Recovering high-quality bacterial genomes from cross-contaminated cultures: a case study of marine Vibrio campbellii
BMC Genomics (2024)
-
Metagenomic exploration of Andaman region of the Indian Ocean
Scientific Reports (2024)
-
High-throughput identification and quantification of bacterial cells in the microbiota based on 16S rRNA sequencing with single-base accuracy using BarBIQ
Nature Protocols (2024)
-
Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont
npj Biofilms and Microbiomes (2024)
-
The core mangrove microbiome reveals shared taxa potentially involved in nutrient cycling and promoting host survival
Environmental Microbiome (2023)