Abstract
Reporting specific modelling methods and metadata is essential to the reproducibility of ecological studies, yet guidelines rarely exist regarding what information should be noted. Here, we address this issue for ecological niche modelling or species distribution modelling, a rapidly developing toolset in ecology used across many aspects of biodiversity science. Our quantitative review of the recent literature reveals a general lack of sufficient information to fully reproduce the work. Over two-thirds of the examined studies neglected to report the version or access date of the underlying data, and only half reported model parameters. To address this problem, we propose adopting a checklist to guide studies in reporting at least the minimum information necessary for ecological niche modelling reproducibility, offering a straightforward way to balance efficiency and accuracy. We encourage the ecological niche modelling community, as well as journal reviewers and editors, to utilize and further develop this framework to facilitate and improve the reproducibility of future work. The proposed checklist framework is generalizable to other areas of ecology, especially those utilizing biodiversity data, environmental data and statistical modelling, and could also be adopted by a broader array of disciplines.
Similar content being viewed by others
Main
Science is facing a reproducibility crisis. A recent Nature survey of 1,576 researchers from various disciplines found that more than 70% of researchers were unable to reproduce research by others, and 50% were not even able to reproduce their own results1. Indeed, the issue of reproducibility has been raised across many fields of science. For instance, the estimates of non-reproducible studies are as high as 89% in cancer research2 and 65% in drug research3, and even high-profile, ‘landmark’ studies are not free of reproducibility issues4. New scientific research builds on previous efforts, allowing methods for testing hypotheses to evolve continually5. Therefore, research results must be communicated with enough context, detail and circumstance to allow correct interpretation, understanding and, whenever possible, reproduction. Reproducibility is a cornerstone of the scientific process and must be emphasized in scientific reports and publications. Although best-practice guidelines have been published and adopted for areas such as computer science6 and clinical research7,8, for various reasons, guidelines for ensuring reproducibility are still largely absent in many (even large) research communities.
Along these lines, the issue of reproducibility may be especially difficult to address in ecology, given the less-controlled aspects of many studies (for example, natural community surveys, field experiments). The issue of reproducibility has been noted only recently in ecology9,10, but is likely prominent11,12. Because ecological studies often encompass uncontrollable or unaccountable factors13, it is especially important to report in detail the circumstances and methods that apply. Furthermore, ecological studies often depend on statistical models, such that reporting specific modelling methods and decisions and how they are intended to reflect biological knowledge or assumptions holds particular importance for reproducibility in ecology14,15. More than ever before, it has become critical to report these aspects, as the data and analytical tools underlying ecological studies are accumulating and evolving at an unprecedented rate in the age of big data16; ecological niche modelling (ENM) is a prominent example.
Ecological niche modelling
Also known as species distribution modelling (SDM)17,18,19, ENM uses associations between known occurrences of species and environmental conditions to estimate species’ potential geographic distributions. Although ENM and SDM are often used interchangeably in the literature20, ENM typically has a stronger focus on estimating parameters of fundamental ecological niches, whereas SDM is more focused on geographic distributions of species. ENM is widely applied across many aspects of ecology and evolution, and is increasingly incorporated in decision-making regarding land use and conservation21. ENM studies are proliferating rapidly; in particular, a popular ENM algorithm, Maxent22, has been cited in tens of thousands of research papers in the past decade alone. Though methods and assumptions in these studies vary greatly, to our knowledge, no evaluation of reproducibility of ENM or SDM studies has been conducted to date (but see ref. 21 for scoring key model aspects for biodiversity assessments). Furthermore, no guidelines on reporting essential modelling parameters exist, hindering accurate evaluation (for example, scoring21) of model methodology and reuse of published research. It is concerning that such a fast-growing and fast-evolving body of literature lacks assessment and guidelines for reproducibility.
Typically, ENM analyses take biodiversity data and environmental data (such as point observations of a species and climate) as input and use correlative or machine-learning methods to quantify underlying relationships, which then are used in making spatial predictions. This typical workflow of ENM — obtaining and processing data, model calibration, model transfer and evaluation — is shared widely across disciplines that rely on statistical models. Therefore, the fast development, broad use and application, and existence of a rather established workflow for ENM makes it an excellent and representative example to tackle the challenges of reproducibility. Here, we assess the reproducibility of ENM studies via a comprehensive literature review and introduce a checklist to facilitate reproducibility of ENMs that can be extended to other areas of ecological research or other disciplines.
A checklist for ecological niche modelling
Although the role of ‘methods’ sections of scientific publications is to provide information that makes the study replicable, they are often highly condensed and lacking details needed for reproducibility, owing in large part to space limitations in journals. What is needed is a standardized format for reporting the full suite of details that comprise the critical information to ensure reproducibility. Therefore, a compendium of crucial parameters and qualities — in effect a metadata standard for ecological niche models — would be highly useful. A metadata standard establishes a common use and understanding through defining a series of attributes and standardized terminology to describe them. Such standards have been applied in various fields, such as GeoTIFF for spatial rasters23 and Darwin Core and Humboldt Core for biodiversity data24,25. A metadata standard can provide a straightforward way to balance efficiency and accuracy in facilitating research reproducibility26 in ENM, as well as scientific studies in general27,28,29.
Here we present a checklist for ENM, to demonstrate how to define general and flexible reproducibility standards that can be used across a wide range of sub-fields of ecology. We compiled a list of essential elements required to reproduce ENM results based on the literature to date, and organized the elements into four major topics: (A) occurrence data collection and processing, (B) environmental data collection and processing, (C) model calibration and (D) model transfer and evaluation (labels correspond to elements in Table 1). We justify the design of the checklist briefly, and provide detailed definitions, examples of reporting for each element, and related literature, in Table 1. We do not distinguish the relative importance among the checklist elements, as all are necessary to assure full reproducibility. We provide a template of the checklist for easier use (Supplementary Table 1). We envision this checklist as a dynamic entity that will continue to be developed and refined by the ENM/SDM community to keep pace with the state of the art in the field. We also provide access to the checklist on Github, as an open-source project where users can comment and suggest changes (https://github.com/shandongfx/ENMchecklist or https://doi.org/10.5281/zenodo.3257732).
Occurrence data (A)
Across many fields, online databases are growing and changing rapidly30, such that reporting data versions or providing complete datasets used in analyses is crucial to reproducibility. Occurrence data are increasingly available owing to mass digitization of museum specimens and increased interest and participation in observational data collection by citizen scientists31. Because the quality of occurrence data can vary significantly among data sources, data types and taxa32,33,34, it is vital to record data curation details to assure consistent quality and accuracy. The first attribute to report is the source of the data (A1; labels correspond to elements in Table 1 hereafter). If the occurrence data were the result of an online database query, the Digital Object Identifier (DOI), query and download date, or the version of a database must also be reported (A2), as online biodiversity data are accumulating rapidly and these data are often edited, corrected, improved or excluded over time35,36,37. The final dataset (that is, after editing and quality control), with the exception of sensitive information (for example, specific locations of endangered taxa), should be deposited in a data archive when reserving rights allow it, thereby assuring reproducibility in case of changes to the original data source.
Whenever available, the ‘basis of record’ (A3) as used in Darwin Core, a community-developed standard for sharing biodiversity data24, should be reported. This field describes how records were originally collected, and thus can indicate different levels of quality and different auxiliary information available. For instance, ‘MachineObservation’ via automated identification may be more prone to error compared with a ‘PreservedSpecimen’ collected and identified by an expert and deposited in a museum. Further, with a deposited specimen and catalogue number, researchers have the opportunity to examine the specimen physically to verify the identification38,39, whereas an observation may not be verifiable. Spatial uncertainty (see A6-3) can vary with the type of occurrence data, as well as the time when the data were collected. For example, coordinates associated with older ‘PreservedSpecimens’ are usually georeferenced from descriptions of administrative units (for example, township, county or country), thus involving higher spatial uncertainty, whereas coordinates linked to recent ‘HumanObservations’ may have been directly reported from GPS devices, making them more accurate. Information regarding the uncertainty of occurrences can also facilitate evaluation of whether the spatial resolution of environmental data utilized is appropriate (see B3). The spatial uncertainty in biodiversity data has long been recognized40,41, though the quantification of such uncertainty has not been implemented systematically at large scale (thus A6-3 was excluded from our literature review; see below); this task could be facilitated by recently developed informatics tools42,43.
Increasingly, ecological research uses data from large-scale data aggregators (for example, the Global Biodiversity Information Facility (GBIF)). As with many sciences relying on observational, rather than design-based data collection, biodiversity data used in ENM have generally not been collected explicitly for this purpose. Thus, the spatial and temporal attributes of occurrences, and how they have been parsed or filtered in preparation for modelling, are essential details required to model ecological niches adequately17. Checking the extent of occurrences (A4) against expert-defined distributions (for example, regional floras) may reduce errors in identification or data transcription. Underrepresentation of the known distribution may suggest inadequate or biased sampling of occurrences, whereas spatial outliers may represent recent range expansion44,45, occasional or vagrant occurrences46, sink populations47, or errors of identification or georeferencing. The collection date of occurrence records may influence spatial accuracy; in general, records from before the 1980s will lack precise point location data (that is, GPS coordinates) and are often georeferenced by hand from locality descriptions and with less precision33. Also, because environments change over time (for example, seasonal change, climate change, land-use changes), the temporal range of the occurrence data (A5) must be specified to connect it appropriately to the temporal dimension of environmental conditions48. Often, occurrence data are processed before modelling (A6). Common procedures include removing duplicate coordinates, excluding spatial and/or environmental outliers, and eliminating records with high spatial uncertainty43 or erroneous coordinates49. Additionally, scholars have proposed various ways to address the well-known issues of sampling bias50,51,52 and spatial autocorrelation53, often by imposing distance-based filters on occurrence data or incorporating spatial structure as a component in the modelling process54 (A7).
Environmental data (B)
Similar to occurrence data, sources for environmental data are numerous, and data often require processing before inclusion in ENM analyses. The source (B1), and database query/download date or version of the database must be reported (B2), as environmental data may be updated periodically (for example, WorldClim55,56) or may accumulate new data regularly through time (for example, PRISM57). Such information is also important for environmental variables derived from remotely sensed data (such as MODIS, Landsat). For example, NASA conducts regular quality assessments of MODIS data products and reprocesses data that may have been influenced by algorithm or calibration issues58.
The spatial resolution of the environmental variables used (B3) can affect ENM results, as different ecological processes occur at different spatial scales59. It has been hypothesized that at broad scales, abiotic conditions have a more dominant role in determining species’ distributions than biotic conditions60,61, though increasing numbers of reported exceptions suggest that this pattern is context dependent62,63. In practice, using different spatial resolutions of environmental variables can produce different results64,65,66. Reporting the spatial resolution of environmental variables can also facilitate checking the match or mismatch with the spatial uncertainty of occurrences, given that coordinates are at times georeferenced from county centroids at coarse resolution33. In addition to reporting the spatial resolution used for modelling, aggregation or disaggregation methods used to align the spatial resolutions of variables (for example, if they came from different data providers) should also be reported.
Providing the temporal range covered by the environmental variables (B4) is important for two reasons67,68. First, shorter temporal ranges can capture finer variation of environments (for example, extremes of daily temperature69), whereas longer temporal ranges capture longer-term trends in environmental conditions (for example, temperature seasonality). Second, it is helpful to evaluate how the temporal range of environmental data relates to the temporal range of occurrence data. For instance, associating occurrence data with environmental data from completely different time periods (for example, Last Glacial Maximum versus present) could be problematic, though the environmental data may need to include time lags to correspond to the life history of particular species70. The same reporting should be applied to information on future or past environments, as appropriate (D9–12). Similarly, the details of methods for processing and resampling of environmental data in temporal dimensions should also be reported.
Model calibration (C)
Typically, an ENM study first has to determine the geographic domain of interest (C1). Delimitation of the domain requires both ecological and practical justification, such as focusing on areas that have been accessible to a species71,72, and areas that have been sampled. Many ENM algorithms make use of background points22 that represent environmental conditions contrasting those known to be occupied by the taxa of interest. Several aspects of background point selection can influence model outcomes, including the number of points (C2)73,74 and the algorithms used to select these points75,76 (C3).
The suite of environmental predictors that are used in ENM should be directly relevant to a species’ distributional ecology19, and the rationale for selecting those variables should be transparent (C4). However, as mechanistic relationships are often unknown, justification of variable selection procedures is necessary. Further, collinearity of environmental variables, a well-recognized issue in regression models, affects parameter estimation during model calibration77; one common strategy is to remove highly correlated environmental variable pairs following rule-of-thumb thresholds (for example, |r| > 0.4 or 0.7)77,78. Selecting one variable from a pair of variables can be subjective (for example, based on expert knowledge), objective (for example, using variable contribution to model fit79) or random; hence justification is required to ensure accurate interpretation and reproducibility of variable selection.
The version of the ENM software or algorithm used (C5 and C6) also needs to be provided, as these tools are often updated80 to include bug fixes or revised default settings. For instance, the default transformation method of Maxent raw output was changed from ‘logistic’ to ‘cloglog’ between versions 3.3 and 3.480. Dependent libraries for coded algorithms may change over time as well.
Parameterizations or model settings and their justification (C7) are important to understanding how they may affect predictions. Examples of these settings include features and regularization values in Maxent81,82, covariate formulas for regression-based models, link functions in generalized linear models (GLMs)83, learning rate and maximum complexity in boosted regression trees (BRTs)84, and optimizer values in generalized additive models 85. In practice, authors often use the default settings provided by the software or algorithm utilized, which may or may not yield robust models72,82,86, wheareas in other cases, authors fine-tune parameters to get best model performance81,87.
Model transfer and evaluation (D)
Understanding model performance requires model evaluation (D1). A first step is that of assessing model precision and significance — that is, whether the model can correctly predict independent presence (or absence) data and whether the model prediction is better than null expectations. Commonly used indices that measure model performance can be either threshold-independent (D2; for example, area under the receiver operating characteristic curve or ROC AUC88), or threshold-dependent (for example, partial ROC89, true skill statistic or TSS, sensitivity and specificity90); the latter approaches require reporting of thresholds and how they were derived. In addition to model accuracy, information criterion-based indices should be reported if they were used to select among competing models based on predictive performance and model complexity or used to generate ensembles of models. Authors should report whether and how data were partitioned to calculate the evaluation indices (D3), if genuinely independent testing data (that is, different sources and methods of collection) were not available. Common approaches include random partitioning of occurrence datasets into training and testing (for example, the default in Maxent); among other methods, partitioning based on structured blocks (for example, separating occurrences into spatial blocks) is expected to assess model transferability better81,91. Given the variety of options regarding data separation, it is important to specify methods used to ensure better reproducibility.
Once a model is calibrated, it may then be transferred or projected onto another landscape or time. Generally, these predictions are initially continuous (D4) and sometimes are subsequently transformed into binary predictions using a particular threshold (D5). Researchers have proposed different ways of thresholding92,93 for different purposes and under varied assumptions, so these choices need to be reported.
Transferring a model across space and/or time may lead to extrapolation if the projected environments are novel relative to training environments. Several studies have found that environmental novelty48,94,95 (D6) and collinearity shift (D7; changes of collinearity structure of covariates77,96) reduce predictive performance, and recommended quantifying the novelty of the projected environments and the collinearity shift between the calibrated and projected environments96,97. Further, different algorithms use different strategies to extrapolate (clamping, truncation, extrapolation94,98); for example, the default clamping function in Maxent uses the marginal values in the calibration area as the prediction for more extreme conditions in transfer areas22.
Assessing the state of reproducibility in ENM research
To assess the state of reproducibility in ENM research in the context of our proposed checklist, we reviewed current (2017–2018) ENM literature in eight widely read ecology and evolution journals: Global Ecology and Biogeography; Diversity and Distributions; Journal of Biogeography; Evolution; Evolutionary Applications; Molecular Phylogenetics and Evolution; Molecular Biology and Evolution; and Systematic Biology. Additional details of our review criteria are provided in Appendix 1, Supplementary Fig. 1, and Supplementary Tables 2 and 3.
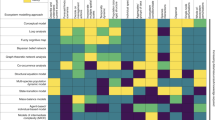
Inclusion of elements of the checklist (32 in total) varied widely, ranging from fully reported (100%; C5 algorithm name) to not reported at all (0%; D7 collinearity shift), though documentation of the importance of this latter element is still limited in the literature77,96. Completeness of information across the checklist also varied among papers, ranging from 24% to 89%, averaging 54% (s.d. = 13%) of checklist elements reported in a given paper (Fig. 1).
Most studies (93%) fully reported sources of occurrence data (A1), but the date of access or version of the data source (A2) was included in only 22% of papers reviewed, and the basis of these records (A3) was described clearly in only 48% of papers. A relatively high number of papers (67%) reported the spatial extent (A4) of the occurrence data, but the temporal range (A5) was mentioned less frequently (26%). Few papers gave details of occurrence data processing, ranging between 18 and 35% in elements A6 and A7.
Although most papers we reviewed reported the source of environmental data (B1), they largely did not include download date or version of the data source: only 27% of papers reported such information for model training (B2) and only 23% for environmental data in model transfer (D10). The spatial resolution and the method of resampling layers with different spatial resolutions (B3) were generally reported (82%), although the temporal range (B4) was less frequently reported (42%). The pattern was opposite for environmental layers for model transfer: temporal range (D12) was almost always reported (94%) but spatial resolution (D11) was less frequently reported (72%).
Only 32% of papers fully reported information regarding modelling domain (C1–3). A high percentage of papers reported the variable selection procedure (C4; 70%). The ENM algorithm or software (C5) was always reported, though less frequently for the corresponding version (C6; 59%). In general, less than half of papers fully disclosed parameters for algorithms (C7; 45%).
Although model evaluation is critical for modelling studies, not all papers (90%) presented information pertaining to model evaluation (D1). Surprisingly, less than half adequately reported how the evaluation dataset was generated (D3; 39%) or mentioned specific values for threshold-dependent evaluation indices (D2; 36%). For model predictions generated, 51% of papers adequately specified output format or acknowledged that default settings were used. Among the papers that converted continuous predictions to binary, 92% specified the adopted threshold. When transferring model to different times and/or regions, few of the papers specified the extrapolation strategy (D8; 36%). The novelty of projected environments (D6) was rarely evaluated (8%).
Lessons from ecological niche modelling
Reproducibility of scientific studies has been under major scrutiny in recent years, and numerous high-profile studies have been found to be irreproducible, in large part because current reporting and publishing practices do not provide sufficient information regarding the methodologies, decisions and assumptions involved. Despite being based on a relatively recently developed toolset, ENM is no exception. For thorough evaluations of proper use of ENM applications (for example, use of ENMs in biodiversity assessment21), a detailed and standardized description of the methods must be provided. The checklist presented here includes the bare minimum of categories and elements necessary to evaluate and replicate ENM analyses. However, the details reported in recent publications varied greatly: on average, papers in our review included only 54% of checklist items, a generally incomplete set of information for reproducibility. This shortcoming may reflect a lack of community expectations on model reporting, or even unawareness of alternative options and underlying caveats in the modelling workflow. We highlight several key areas that were particularly deficient in reporting, and thus need attention to make ENM studies reproducible (Box 1).
Improving reproducibility with software solutions
The rapid development of ENM can be attributed at least in part to increased access to relevant data; with such development, informatics tools offer one route by which to improve reproducibility99,100. Such tools include data management plans101, standardized metadata102,103, programming language resources to record data analysis steps (for example, R and rmarkdown) and version-control tools (for example, GitHub). Open-source programming languages such as R have allowed for development of packages specifically designed for managing and processing large datasets in preparation for analysis. Exemplary packages include biogeo, which directly detects, corrects, and assesses occurrence data quality42, and geoknife, a package designed specifically for United States Geological Survey gridded dataset management104. Other packages help users to create reproducible workflows, such as zoon105, nicheA106 kuenm86, and Wallace107. In particular, the package Wallace provides a graphical user interface to build reproducible workflows, from data download to model output107. Borregaard and Hart11 described how the use of these new software tools is facilitating ecological research that is both robust and transparent, and thus reproducible. The functionality of the software solutions, however, depends on developers monitoring changes in data, modelling algorithms and the software platforms (for example, R), to avoid incompatibility issues. As such, authors should report software versions for all such solutions to ensure reproducibility.
Implications for other fields
The design of the checklist presented here is based on a typical ENM workflow, involving steps of obtaining and processing data, and model calibration, transfer and evaluation. We emphasized reporting data origin and metadata; crucial steps in data processing, modelling decisions and model evaluation; and potential caveats in model transfer. Those concepts and principles are generalizable to other disciplines. Further, the specifics of the checklist that we have proposed for ENM studies could be readily generalized to be adopted by other fields, especially those that involve biological data, environmental data and statistical modelling.
Researchers have proposed similar solutions in other fields, such as climate change research29; however, to our knowledge, our checklist takes additional steps in refining the methodology workflow and is therefore more comprehensive. For example, information pertaining to occurrence data (data source, spatial and temporal range, and data cleaning procedures) can be generalized to other studies that rely on digitized biodiversity data and other categories of ‘big data’. The information regarding environmental data necessary to reproduce studies is similar across biological research, such as in studies of relationships between species richness and environmental gradients108. The modelling algorithm details in the checklist are applicable to other studies that use statistical models, such as linear regression models of abundance as a response to resource availability. The elements of model extrapolation (environmental novelty and collinearity shift) are also common issues for modelling practices that involve forecasting, for example, predictions of biodiversity or community changes under global change. In addition to these generally applicable elements, the checklist can easily be extended to incorporate information particular to a field.
Although the methods sections of most scientific publications lack the formal standardization needed for reproducibility and their length is frequently influenced by journal space limitations, the checklist approach can provide greater detail to ensure repeatability. The usual methods section, combined with a standardized checklist, will make papers easier to review and replicate. Other disciplines can and should design comparable checklists with similar concepts and levels of detail.
Closing remarks
ENM is increasingly used in ecological studies and incorporated into conservation decisions. Our literature review revealed numerous gaps that undermine reproducibility of these studies. We recommend researchers developing ENM studies in the future to consider our checklist, extend and adjust it to meet study needs, with particular focus on elements that are commonly neglected (Table 1), and include this more structured metadata in publications (see checklist template in Supplementary Table 1). This checklist provides an important tool for both understanding and replicating previous studies, and also provides editors and reviewers with an efficient way to gauge and promote ENM reproducibility29. As a general metadata framework linking observational data and statistical modelling, our checklist provides a starting point for adopting similar standards in other fields, both within and beyond ecology that rely on these methods.
Data availability
The checklist for ENM can be downloaded from Supplementary Table 1 and is available as an open-source project where users can comment and suggest changes (https://github.com/shandongfx/ENMchecklist or https://doi.org/10.5281/zenodo.3257732). Details of the literature review are available in the Supplementary Information.
References
Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016).
Begley, C. G. & Ellis, L. M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Mullard, A. Reliability of ‘new drug target’ claims called into question. Nat. Rev. Drug Discov. 10, 643–644 (2011).
Problems with scientific research: how science goes wrong. The Economist https://www.economist.com/leaders/2013/10/21/how-science-goes-wrong (21 October 2013).
Popper, K. Conjectures and Refutations: The Growth of Scientific Knowledge (Routledge, 2014).
Wilson, G. et al. Best practices for scientific computing. PLoS Biol. 12, e1001745 (2014).
Nichols, T. E. et al. Best practices in data analysis and sharing in neuroimaging using MRI. Nat. Neurosci. 20, 299–303 (2017).
Tumor Analysis Best Practices Working Group. Expression profiling: best practices for data generation and interpretation in clinical trials. Nat. Rev. Genet. 5, 229–237 (2004).
Cassey, P. & Blackburn, T. M. Reproducibility and repeatability in ecology. Bioscience 56, 958–959 (2006).
Shapiro, J. T. & Báldi, A. Lost locations and the (ir)repeatability of ecological studies. Front. Ecol. Environ. 10, 235–236 (2012).
Borregaard, M. K. & Hart, E. M. Towards a more reproducible ecology. Ecography 39, 349–353 (2016).
Schnitzer, S. A. & Carson, W. P. Would ecology fail the repeatability test? Bioscience 66, 98–99 (2016).
Milcu, A. et al. Genotypic variability enhances the reproducibility of an ecological study. Nat. Ecol. Evol. 2, 279–287 (2018).
Nekrutenko, A. & Taylor, J. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet. 13, 667–672 (2012).
Hunter, P. The reproducibility ‘crisis’: reaction to replication crisis should not stifle innovation. EMBO Rep. 18, 1493–1496 (2017).
Hampton, S. E. et al. Big data and the future of ecology. Front. Ecol. Environ. 11, 156–162 (2013).
Peterson, A. T. et al. Ecological Niches and Geographic Distributions (Princeton Univ. Press, 2011).
Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction (Cambridge Univ. Press, 2010).
Guisan, A. & Zimmermann, N. E. Predictive habitat distribution models in ecology. Ecol. Model. 135, 147–186 (2000).
Peterson, A. T. & Soberón, J. Species distribution modeling and ecological niche modeling: getting the concepts right. Nat. Conservação 10, 102–107 (2012).
Araújo, M. B. et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 5, eaat4858 (2019).
Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006).
Mahammad, S. S. & Ramakrishnan, R. GeoTIFF: a standard image file format for GIS applications. In Map India Conf. 2003 28–31 (2003).
Wieczorek, J. et al. Darwin Core: an evolving community-developed biodiversity data standard. PLoS ONE 7, e29715 (2012).
Guralnick, R., Walls, R. & Jetz, W. Humboldt Core: toward a standardized capture of biological inventories for biodiversity monitoring, modeling and assessment. Ecography 41, 713–725 (2018).
Gad-el-Hak, M. Publish or perish—an ailing enterprise? Phys. Today 57, 61–62 (2004).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017).
Grimm, V. et al. A standard protocol for describing individual-based and agent-based models. Ecol. Model. 198, 115–126 (2006).
Morueta-Holme, N. et al. Best practices for reporting climate data in ecology. Nat. Clim. Change 8, 92–94 (2018).
Michener, W. K. et al. Participatory design of DataONE — enabling cyberinfrastructure for the biological and environmental sciences. Ecol. Inform. 11, 5–15 (2012).
Bonney, R. et al. Citizen science: a developing tool for expanding science knowledge and scientific literacy. Bioscience 59, 977–984 (2009).
Daru, B. H. et al. Widespread sampling biases in herbaria revealed from large-scale digitization. New Phytol. 217, 939–955 (2018).
Park, D. S. & Davis, C. C. Implications and alternatives of assigning climate data to geographical centroids. J. Biogeogr. 44, 2188–2198 (2017).
Meyer, C., Weigelt, P. & Kreft, H. Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19, 992–1006 (2016).
Enquist, B. J., Condit, R., Peet, R. K., Schildhauer, M. & Thiers, B. M. Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. Preprint at https://doi.org/10.7287/peerj.preprints.2615v2 (2016).
Boyle, B. et al. The taxonomic name resolution service: an online tool for automated standardization of plant names. BMC Bioinformatics 14, 16 (2013).
iNaturalist Research-grade Observations (iNaturalist.org, 2018); https://doi.org/10.15468/ab3s5x
Castro, M. C. et al. Reassessment of the hairy long-nosed armadillo ‘Dasypus’ pilosus (Xenarthra, Dasypodidae) and revalidation of the genus Cryptophractus Fitzinger, 1856. Zootaxa 3947, 30–48 (2015).
Park, D. S. & Potter, D. A reciprocal test of Darwin’s naturalization hypothesis in two mediterranean-climate regions. Glob. Ecol. Biogeogr. 24, 1049–1058 (2015).
Guralnick, R. P., Wieczorek, J., Beaman, R. & Hijmans, R. J. & the BioGeomancer Working Group BioGeomancer: automated georeferencing to map the world’s biodiversity data. PLoS Biol. 4, e381 (2006).
Wieczorek, J., Guo, Q. & Hijmans, R. The point-radius method for georeferencing locality descriptions and calculating associated uncertainty. Int. J. Geogr. Inf. Sci. 18, 745–767 (2004).
Robertson, M. P., Visser, V. & Hui, C. Biogeo: an R package for assessing and improving data quality of occurrence record datasets. Ecography 39, 394–401 (2016).
Zizka, A. et al. CoordinateCleaner: standardized cleaning of occurrence records from biological collection databases. Methods Ecol. Evol. 10, 744–751 (2019).
McCracken, G. F. et al. Rapid range expansion of the Brazilian free-tailed bat in the southeastern United States, 2008–2016. J. Mammal. 99, 312–320 (2018).
Taulman, J. F. & Robbins, L. W. Range expansion and distributional limits of the nine-banded armadillo in the United States: an update of Taulman & Robbins (1996). J. Biogeogr. 41, 1626–1630 (2014).
Feng, X. & Papeş, M. Ecological niche modelling confirms potential north-east range expansion of the nine-banded armadillo (Dasypus novemcinctus) in the USA. J. Biogeogr. 42, 803–807 (2015).
Pulliam, H. R. On the relationship between niche and distribution. Ecol. Lett. 3, 349–361 (2000).
Fitzpatrick, M. C. et al. How will climate novelty influence ecological forecasts? Using the Quaternary to assess future reliability. Glob. Ecol. Biogeogr. 24, 3575–3586 (2018).
Belbin, L. et al. Data Quality Task Group 2: tests and assertions. BISS 2, e25608 (2018).
Fourcade, Y., Engler, J. O., Rödder, D. & Secondi, J. Mapping species distributions with MAXENT using a geographically biased sample of presence data: a performance assessment of methods for correcting sampling bias. PLoS ONE 9, e97122 (2014).
Phillips, S. J. et al. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 19, 181–197 (2009).
Merow, C., Allen, J. M., Aiello-Lammens, M. & Silander, J. A. Jr Improving niche and range estimates with Maxent and point process models by integrating spatially explicit information. Glob. Ecol. Biogeogr. 25, 1022–1036 (2016).
Dormann, C. F. et al. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628 (2007).
Latimer, A. M., Banerjee, S., Sang, H. Jr, Mosher, E. S. & Silander, J. A. Jr Hierarchical models facilitate spatial analysis of large data sets: a case study on invasive plant species in the northeastern United States. Ecol. Lett. 12, 144–154 (2009).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
PRISM Gridded Climate Data (PRISM Climate Group, accessed 1 July 2017); http://prism.oregonstate.edu
QA Note: Case #PM_MOD16_17166 (LAADS and DAAC, 2017); https://go.nature.com/2lu5NCw
McGill, B. J. Matters of scale. Science 328, 575–576 (2010).
Soberón, J. & Nakamura, M. Niches and distributional areas: concepts, methods, and assumptions. Proc. Natl Acad. Sci. USA 106, 19644–19650 (2009). (suppl. 2).
Sunday, J. M., Bates, A. E. & Dulvy, N. K. Thermal tolerance and the global redistribution of animals. Nat. Clim. Change 2, 686–690 (2012).
Wisz, M. S. et al. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biol. Rev. 88, 15–30 (2013).
Alexander, J. M., Diez, J. M. & Levine, J. M. Novel competitors shape species’ responses to climate change. Nature 525, 515–518 (2015).
Bradter, U., Kunin, W. E., Altringham, J. D., Thom, T. J. & Benton, T. G. Identifying appropriate spatial scales of predictors in species distribution models with the random forest algorithm. Methods Ecol. Evol. 4, 167–174 (2012).
Song, W., Kim, E., Lee, D., Lee, M. & Jeon, S.-W. The sensitivity of species distribution modeling to scale differences. Ecol. Model. 248, 113–118 (2013).
Connor, T. et al. Effects of grain size and niche breadth on species distribution modeling. Ecography 41, 1270–1282 (2018).
Smeraldo, S. et al. Ignoring seasonal changes in the ecological niche of non-migratory species may lead to biases in potential distribution models: lessons from bats. Biodiv. Conserv. 27, 2425–2441 (2018).
Fernandez, M., Yesson, C., Gannier, A., Miller, P. I. & Azevedo, J. M. N. The importance of temporal resolution for niche modelling in dynamic marine environments. J. Biogeogr. 44, 2816–2827 (2017).
Barve, N., Martin, C., Brunsell, N. A. & Peterson, A. T. The role of physiological optima in shaping the geographic distribution of Spanish moss: physiological optima of Spanish moss. Glob. Ecol. Biogeogr. 23, 633–645 (2014).
Williams, H. M., Willemoes, M. & Thorup, K. A temporally explicit species distribution model for a long distance avian migrant, the common cuckoo. J. Avian Biol. 48, 1624–1636 (2017).
Barve, N. et al. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol. Model. 222, 1810–1819 (2011).
Merow, C., Smith, M. J. & Silander, J. A.Jr. A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36, 1058–1069 (2013).
Barbet-Massin, M., Jiguet, F., Albert, C. H. & Thuiller, W. Selecting pseudo-absences for species distribution models: how, where and how many? Methods Ecol. Evol. 3, 327–338 (2012).
VanDerWal, J., Shoo, L. P., Graham, C. & Williams, S. E. Selecting pseudo-absence data for presence-only distribution modeling: How far should you stray from what you know? Ecol. Model. 220, 589–594 (2009).
Senay, S. D., Worner, S. P. & Ikeda, T. Novel three-step pseudo-absence selection technique for improved species distribution modelling. PLoS ONE 8, e71218 (2013).
Feng, X. & Papeş, M. Can incomplete knowledge of species’ physiology facilitate ecological niche modelling? A case study with virtual species. Divers. Distrib. 23, 1157–1168 (2017).
Dormann, C. F. et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36, 27–46 (2013).
Suzuki, N., Olson, D. H. & Reilly, E. C. Developing landscape habitat models for rare amphibians with small geographic ranges: a case study of Siskiyou Mountains salamanders in the western USA. Biodiv. Conserv. 17, 2197–2218 (2008).
Lee, D. N., Papeş, M. & Van den Bussche, R. A. Present and potential future distribution of common vampire bats in the Americas and the associated risk to cattle. PLoS ONE 7, e42466 (2012).
Phillips, S. J., Anderson, R. P., Dudík, M., Schapire, R. E. & Blair, M. E. Opening the black box: an open-source release of Maxent. Ecography 40, 887–893 (2017).
Muscarella, R. et al. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 5, 1198–1205 (2014).
Phillips, S. J. & Dudík, M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175 (2008).
Elith, J. et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29, 129–151 (2006).
Elith, J., Leathwick, J. R. & Hastie, T. A working guide to boosted regression trees. J. Appl. Ecol. 77, 802–813 (2008).
Lehmann, A., Overton, J. M. & Leathwick, J. R. GRASP: generalized regression analysis and spatial prediction. Ecol. Model. 157, 189–207 (2002).
Cobos, M. E., Peterson, A. T., Osorio-Olvera, L. & Narayani, B. kuenm: An R package for detailed development of Maxent ecological niche models. PeerJ 7, e6281 (2019).
Moreno-Amat, E. et al. Impact of model complexity on cross-temporal transferability in Maxent species distribution models: an assessment using paleobotanical data. Ecol. Model. 312, 308–317 (2015).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988).
Peterson, A. T., Papeş, M. & Soberón, J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol. Model. 213, 63–72 (2008).
Allouche, O., Tsoar, A. & Kadmon, R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS): assessing the accuracy of distribution models. J. Appl. Ecol. 43, 1223–1232 (2006).
Roberts, D. R. et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 40, 913–929 (2017).
Liu, C., Berry, P. M., Dawson, T. P. & Pearson, R. G. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28, 385–393 (2005).
Liu, C., White, M. & Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 40, 778–789 (2013).
Owens, H. L. et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol. Model. 263, 10–18 (2013).
Elith, J., Kearney, M. & Phillips, S. The art of modelling range-shifting species. Methods Ecol. Evol. 1, 330–342 (2010).
Feng, X., Park, D. S., Pandey, R., Liang, Y. & Papeş, M. Collinearity in ecological niche modeling: confusions and challenges. Ecol. Evol. https://doi.org/10.1002/ece3.5555 (2019).
Qiao, H. et al. An evaluation of transferability of ecological niche models. Ecography 42, 521–534 (2019).
Elith, J. & Graham, C. H. Do they? How do they? Why do they differ? On finding reasons for differing performances of species distribution models. Ecography 32, 66–77 (2009).
Soberón, J. & Peterson, A. T. Biodiversity informatics: managing and applying primary biodiversity data. Phil. Trans. R. Soc. Lond. B 359, 689–698 (2004).
Boyd, D. S. & Foody, G. M. An overview of recent remote sensing and GIS based research in ecological informatics. Ecol. Inform. 6, 25–36 (2011).
Michener, W. K. in Ecological Informatics (eds. Recknagel, F. & Michener, W.) 13–26 (Springer, 2018).
Borba, C. & Correa, P. L. P. in Metadata and Semantics Research. MTSR 2015 (eds. Garoufallou, E., Hartley, R. & Gaitanou, P.) 113–118 (Communications in Computer and Information Science Vol. 544, Springer, 2015).
Merow, C. et al. Species’ range model metadata standards: RMMS. Glob. Ecol. Biogeogr. https://doi.org/10.1111/geb.12993 (2019).
Read, J. S. et al. geoknife: reproducible web-processing of large gridded datasets. Ecography 39, 354–360 (2016).
Golding, N. et al. The zoon R package for reproducible and shareable species distribution modelling. Methods Ecol. Evol. 9, 260–268 (2018).
Qiao, H. et al. NicheA: creating virtual species and ecological niches in multivariate environmental scenarios. Ecography 39, 805–813 (2016).
Kass, J. M. et al. Wallace: A flexible platform for reproducible modeling of species niches and distributions built for community expansion. Methods Ecol. Evol. 9, 1151–1156 (2018).
Sandel, B. et al. The influence of Late Quaternary climate-change velocity on species endemism. Science 334, 660–664 (2011).
Bosch, S., Tyberghein, L., Deneudt, K., Hernandez, F. & De Clerck, O. In search of relevant predictors for marine species distribution modelling using the MarineSPEED benchmark dataset. Divers. Distrib. 24, 144–157 (2018).
Franklin, J., Serra-Diaz, J. M., Syphard, A. D. & Regan, H. M. Big data for forecasting the impacts of global change on plant communities. Glob. Ecol. Biogeogr. 26, 6–17 (2017).
McMinn, R. L., Russell, F. L. & Beck, J. B. Demographic structure and genetic variability throughout the distribution of Platte thistle (Cirsium canescens Asteraceae). J. Biogeogr. 44, 375–385 (2017).
Graham, C. H., Ferrier, S., Huettman, F., Moritz, C. & Peterson, A. T. New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol. Evol. 19, 497–503 (2004).
Jetz, W., McPherson, J. M. & Guralnick, R. P. Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159 (2012).
Edwards, T. C. Jr, Cutler, D. R., Zimmermann, N. E., Geiser, L. & Moisen, G. G. Effects of sample survey design on the accuracy of classification tree models in species distribution models. Ecol. Model. 199, 132–141 (2006).
Mammola, S. & Isaia, M. Rapid poleward distributional shifts in the European cave-dwelling Meta spiders under the influence of competition dynamics. J. Biogeogr. 44, 2789–2797 (2017).
Soley-Guardia, M., Radosavljevic, A., Rivera, J. L. & Anderson, R. P. The effect of spatially marginal localities in modelling species niches and distributions. J. Biogeogr. 41, 1390–1401 (2014).
McPherson, J. M., Walter, J. & Rogers, D. J. The effects of species’ range sizes on the accuracy of distribution models: ecological phenomenon or statistical artefact? J. Appl. Ecol. 41, 811–823 (2004).
Phillips, N. D. et al. Applying species distribution modelling to a data poor, pelagic fish complex: the ocean sunfishes. J. Biogeogr. 44, 2176–2187 (2017).
Lee, T. R. C. et al. Ecological diversification of the Australian Coptotermes termites and the evolution of mound building. J. Biogeogr. 44, 1405–1417 (2017).
Boria, R. A., Olson, L. E., Goodman, S. M. & Anderson, R. P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 275, 73–77 (2014).
Ceolin, G. B. & Giehl, E. L. H. A little bit everyday: range size determinants in Arachis (Fabaceae), a dispersal-limited group. J. Biogeogr. 44, 2798–2807 (2017).
Kumar, S., Graham, J., West, A. M. & Evangelista, P. H. Using district-level occurrences in MaxEnt for predicting the invasion potential of an exotic insect pest in India. Comput. Electron. Agric. 103, 55–62 (2014).
Gomes, V. H. F. et al. Species distribution modelling: contrasting presence-only models with plot abundance data. Sci. Rep. 8, 1003 (2018).
Boria, R. A., Olson, L. E., Goodman, S. M. & Anderson, R. P. A single-algorithm ensemble approach to estimating suitability and uncertainty: cross-time projections for four Malagasy tenrecs. Divers. Distrib. 23, 196–208 (2017).
Varela, S., Anderson, R. P., García-Valdés, R. & Fernández-González, F. Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography 33, 1084–1091 (2014).
Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B. & Anderson, R. P. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38, 541–545 (2015).
Hijmans, R. J. Cross-validation of species distribution models: removing spatial sorting bias and calibration with a null model. Ecology 93, 679–688 (2012).
Hortal, J., Valverde, A. J., Gómez, J. F. & Lobo, J. M. Historical bias in biodiversity inventories affects the observed environmental niche of the species. Oikos 117, 847–858 (2008).
Royle, J. A., Nichols, J. D. & Kéry, M. Modelling occurrence and abundance of species when detection is imperfect. Oikos 110, 353–359 (2005).
Segurado, P. & Araújo, M. B. An evaluation of methods for modelling species distributions. J. Biogeogr. 31, 1555–1568 (2004).
Latimer, A. M., Wu, S., Gelfand, A. E. & Silander, J. A. Building statistical models to analyze species distributions. Ecol. Appl. 16, 33–50 (2006).
Record, S., Fitzpatrick, M. C., Finley, A. O., Veloz, S. & Ellison, A. M. Should species distribution models account for spatial autocorrelation? A test of model projections across eight millennia of climate change: Projecting spatial species distribution models. Glob. Ecol. Biogeogr. 22, 760–771 (2013).
Wintle, B. A. & Bardos, D. C. Modeling species-habitat relationships with spatially autocorrelated observation data. Ecol. Appl. 16, 1945–1958 (2006).
Figueiredo, F. O. G. et al. Beyond climate control on species range: the importance of soil data to predict distribution of Amazonian plant species. J. Biogeogr. 45, 190–200 (2018).
Guisan, A., Graham, C. H., Elith, J. & Huettmann, F. Sensitivity of predictive species distribution models to change in grain size. Divers. Distrib. 13, 332–340 (2007).
Sofaer, H. R., Jarnevich, C. S. & Flather, C. H. Misleading prioritizations from modelling range shifts under climate change. Glob. Ecol. Biogeogr. 27, 658–666 (2018).
Cooper, J. C. & Soberón, J. Creating individual accessible area hypotheses improves stacked species distribution model performance. Glob. Ecol. Biogeogr. 27, 156–165 (2018).
Acevedo, P., Jiménez-Valverde, A., Lobo, J. M. & Real, R. Delimiting the geographical background in species distribution modelling. J. Biogeogr. 39, 1383–1390 (2012).
Qiao, H., Escobar, L. E. & Peterson, A. T. Accessible areas in ecological niche comparisons of invasive species: recognized but still overlooked. Sci. Rep. 7, 1213 (2017).
Saupe, E. E. et al. Variation in niche and distribution model performance: the need for a priori assessment of key causal factors. Ecol. Model. 237–238, 11–22 (2012).
Hill, M. P., Gallardo, B. & Terblanche, J. S. A global assessment of climatic niche shifts and human influence in insect invasions. Glob. Ecol. Biogeogr. 26, 679–689 (2017).
Renner, I. W. & Warton, D. I. Equivalence of Maxent and Poisson point process models for species distribution modeling in ecology. Biometrics 69, 274–281 (2013).
Scofield, R. P. et al. The origin and phylogenetic relationships of the New Zealand ravens. Mol. Phylogen. Evol. 106, 136–143 (2017).
Iturbide, M. et al. A framework for species distribution modelling with improved pseudo-absence generation. Ecol. Model. 312, 166–174 (2015).
Hertzog, L. R., Besnard, A. & Jay-Robert, P. Field validation shows bias-corrected pseudo-absence selection is the best method for predictive species-distribution modelling. Divers. Distrib. 20, 1403–1413 (2014).
Warton, D. I. & Shepherd, L. C. Poisson point process models solve the ‘pseudo-absence problem’ for presence-only data in ecology. Ann. Appl. Stat. 4, 1383–1402 (2010).
Beyer, H. L. et al. The interpretation of habitat preference metrics under use-availability designs. Phil. Trans. R. Soc. Lond. B 365, 2245–2254 (2010).
Petitpierre, B., Broennimann, O., Kueffer, C., Daehler, C. & Guisan, A. Selecting predictors to maximize the transferability of species distribution models: lessons from cross-continental plant invasions. Glob. Ecol. Biogeogr. 26, 275–287 (2017).
Sochor, M., Šarhanová, P., Pfanzelt, S. & Trávníček, B. Is evolution of apomicts driven by the phylogeography of the sexual ancestor? Insights from European and caucasian brambles (Rubus, Rosaceae). J. Biogeogr. 44, 2717–2728 (2017).
Derville, S., Torres, L. G., Iovan, C. & Garrigue, C. Finding the right fit: Comparative cetacean distribution models using multiple data sources and statistical approaches. Divers. Distrib. 24, 1657–1673 (2018).
Guisan, A., Thuiller, W. & Zimmermann, N. E. Habitat Suitability and Distribution Models: With Applications in R (Cambridge Univ. Press, 2017).
Qiao, H., Soberón, J. & Peterson, A. T. No silver bullets in correlative ecological niche modelling: insights from testing among many potential algorithms for niche estimation. Methods Ecol. Evol. 6, 1126–1136 (2015).
Herrera, J. P. et al. Estimating the population size of lemurs based on their mutualistic food trees. J. Biogeogr. 45, 2546–2563 (2018).
Warren, D. L. & Seifert, S. N. Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecol. Appl. 21, 335–342 (2011).
Elith, J., Leathwick, J. R. & Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 77, 802–813 (2008).
Anderson, R. P. & Gonzalez, I. Species-specific tuning increases robustness to sampling bias in models of species distributions: an implementation with Maxent. Ecol. Model. 222, 2796–2811 (2011).
Guillera-Arroita, G. et al. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 24, 276–292 (2015).
Martínez-Gutiérrez, P. G., Martínez-Meyer, E., Palomares, F. & Fernández, N. Niche centrality and human influence predict rangewide variation in population abundance of a widespread mammal: the collared peccary (Pecari tajacu). Divers. Distrib. 24, 103–115 (2018).
Swets, J. A. Measuring the accuracy of diagnostic systems. Science 240, 1285–1293 (1988).
Lobo, J. M., Jiménez-Valverde, A. & Real, R. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145–151 (2008).
Liu, C., White, M. & Newell, G. Measuring and comparing the accuracy of species distribution models with presence-absence data. Ecography 34, 232–243 (2011).
Bahn, V. & Mcgill, B. J. Testing the predictive performance of distribution models. Oikos 122, 321–331 (2012).
Boyce, M. S., Vernier, P. R., Nielsen, S. E. & Schmiegelow, F. Evaluating resource selection functions. Ecol. Model. 157, 281–300 (2002).
Lawson, C. R., Hodgson, J. A., Wilson, R. J. & Richards, S. A. Prevalence, thresholds and the performance of presence-absence models. Methods Ecol. Evol. 5, 54–64 (2013).
Fielding, A. H. & Bell, J. F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49 (1997).
Liu, C., Newell, G. & White, M. On the selection of thresholds for predicting species occurrence with presence-only data. Ecol. Evol. 6, 337–348 (2015).
Johnston, M. R., Elmore, A. J., Mokany, K., Lisk, M. & Fitzpatrick, M. C. Field-measured variables outperform derived alternatives in Maryland stream biodiversity models. Divers. Distrib. 23, 1054–1066 (2017).
Hastie, T., Tibshirani, R. & Friedman, J. H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer, 2009).
Elith, J. et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17, 43–57 (2011).
Royle, J. A., Chandler, R. B., Yackulic, C. & Nichols, J. D. Likelihood analysis of species occurrence probability from presence only data for modelling species distributions. Methods Ecol. Evol. 3, 545–554 (2012).
Bartoleti, L. F. M. et al. Phylogeography of the dry vegetation endemic species Nephila sexpunctata (Araneae: Araneidae) suggests recent expansion of the Neotropical Dry Diagonal. J. Biogeogr. 44, 2007–2020 (2017).
Pearson, R. G., Raxworthy, C. J., Nakamura, M. & Peterson, A. T. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J. Biogeogr. 34, 102–117 (2006).
Di Febbraro, M. et al. Does the jack of all trades fare best? Survival and niche width in Late Pleistocene megafauna. J. Biogeogr. 44, 2828–2838 (2017).
Peterson, A. T., Papeş, M. & Eaton, M. Transferability and model evaluation in ecological niche modeling: a comparison of GARP and Maxent. Ecography 30, 550–560 (2007).
Randin, C. F. et al. Are niche-based species distribution models transferable in space? J. Biogeogr. 33, 1689–1703 (2006).
Feng, X., Lin, C., Qiao, H. & Ji, L. Assessment of climatically suitable area for Syrmaticus reevesii under climate change. Endanger. Species Res. 28, 19–31 (2015).
Braunisch, V. et al. Selecting from correlated climate variables: a major source of uncertainty for predicting species distributions under climate change. Ecography 36, 971–983 (2013).
Zurell, D., Elith, J. & Schröder, B. Predicting to new environments: tools for visualizing model behaviour and impacts on mapped distributions. Divers. Distrib. 18, 628–634 (2012).
Matsumoto, M. & Nishimura, T. Mersenne twister: a 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans. Model. Comput. Sim. 8, 3–30 (1998).
Bak, P. & Sneppen, K. Punctuated equilibrium and criticality in a simple model of evolution. Phys. Rev. Lett. 71, 4083–4086 (1993).
Newman, M. E. J., Watts, D. J. & Strogatz, S. H. Random graph models of social networks. Proc. Natl Acad. Sci. USA 99, 2566–2572 (2002). (suppl. 1).
Citation guidelines. GBIF https://www.gbif.org/citation-guidelines (2018).
Peterson, A. T., Papeş, M. & Soberón, J. Mechanistic and correlative models of ecological niches. Eur. J. Ecol. 1, 28–38 (2015).
Acknowledgements
X.F., C.W., A.T.P. and M.P. acknowledge the National Institute for Mathematical and Biological Synthesis (NIMBioS) for facilitating initial discussions of this work. X.F. and D.S.P. acknowledge support from The University of Arizona Office of Research, Discovery, and Innovation, Institute of the Environment, the Udall Center for Studies in Public Policy, and the College of Science on the postdoctoral cluster initiative—Bridging Biodiversity and Conservation Science. C.M. acknowledges funding from NSF grants DBI-1913673 and DBI-1661510.
Author information
Authors and Affiliations
Contributions
X.F. and D.S.P. conceived the idea that was refined by C.W., A.T.P., C.M. and M.P.; X.F., D.S.P. and C.W. conducted the literature review; X.F. conducted the analyses and drafted the manuscript. All authors contributed to the revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Appendix 1, Supplementary Fig. 1 and Table 2.
Supplementary Table 1
Template of ecological niche modelling (ENM) checklist for users to fill.
Supplementary Table 3
List of papers used in the literature review.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, X., Park, D.S., Walker, C. et al. A checklist for maximizing reproducibility of ecological niche models. Nat Ecol Evol 3, 1382–1395 (2019). https://doi.org/10.1038/s41559-019-0972-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0972-5
This article is cited by
-
Ecological niche modelling of Indigofera oblongifolia (Forssk.): a global machine learning assessment using climatic and non-climatic predictors
Discover Environment (2024)
-
A systematic review on the potential impact of future climate change on India’s biodiversity using species distribution model (SDM) studies: trends, and data gaps
Biodiversity and Conservation (2024)
-
Predicted changes in distribution and richness of wild edible plants under climate change scenarios in northwestern Kenya
Regional Environmental Change (2024)
-
Global distribution of the invasive apple snail Pomacea canaliculata: analyzing possible shifts in climatic niche between native and invaded ranges and future spread
Aquatic Sciences (2024)
-
Transmission risk of Oropouche fever across the Americas
Infectious Diseases of Poverty (2023)