Abstract
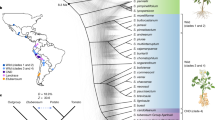
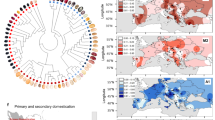
Potato, one of the most important staple crops, originates from the highlands of the equatorial Andes. There, potatoes propagate vegetatively via tubers under short days, constant throughout the year. After their introduction to Europe in the sixteenth century, potatoes adapted to a shorter growing season and to tuber formation under long days. Here, we traced the demographic and adaptive history of potato introduction to Europe. To this end, we sequenced 88 individuals that comprise landraces, modern cultivars and historical herbarium samples, including specimens collected by Darwin during the voyage of the Beagle. Our findings show that European potatoes collected during the period 1650–1750 were closely related to Andean landraces. After their introduction to Europe, potatoes admixed with Chilean genotypes. We identified candidate genes putatively involved in long-day pre-adaptation, and showed that the 1650–1750 European individuals were not long-day adapted through previously described allelic variants of the CYCLING DOF FACTOR1 gene. Such allelic variants were detected in Europe during the nineteenth century. Our study highlights the power of combining contemporary and historical genomes to understand the complex evolutionary history of crop adaptation to new environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data generated in this study are available in the European Nucleotide Archive (ENA) under project Nos. PRJEB31013 and PRJEB31171 for UDG-treated and non-UDG-treated libraries, respectively.
References
Spooner, D. M., McLean, K., Ramsay, G., Waugh, R. & Bryan, G. J. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl Acad. Sci. USA 102, 14694–14699 (2005).
Hawkes, J. G. & Francisco-Ortega, J. The early history of the potato in Europe. Euphytica 70, 1–7 (1993).
Ames, M. & Spooner, D. M. DNA from herbarium specimens settles a controversy about origins of the European potato. Am. J. Bot. 95, 252–257 (2008).
Mcneill, W. H. How the potato changed the world’s history. Soc. Res. 66, 67–83 (1999).
Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 418, 700–707 (2002).
Allaby, R. G. et al. Using archaeogenomic and computational approaches to unravel the history of local adaptation in crops. Philos. Trans. R. Soc. Lond. B 370, 20130377 (2015).
Shennan, S. et al. Regional population collapse followed initial agriculture booms in mid-Holocene Europe. Nat. Commun. 4, 2486 (2013).
Colledge, S., Conolly, J. & Shennan, S. The evolution of neolithic farming from SW asian origins to NW european limits. Eur. J. Archaeol. 8, 137–156 (2016).
Fuller, D. Q. & Allaby, R. G. in Fruit Development and Seed Dispersal (ed. Ostergaard, L.) 238–295 (Blackwell, 2009).
Jackson, S. D. Multiple signaling pathways control tuber induction in potato. Plant Physiol. 119, 1–8 (1999).
Kloosterman, B. et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250 (2013).
Spooner, D., Jansky, S., Clausen, A., del Rosario Herrera, M. & Ghislain, M. The enigma of Solanum maglia in the origin of the chilean cultivated potato, Solanum tuberosum Chilotanum group. Econ. Bot. 66, 12–21 (2012).
Ghislain, M., Núñez, J., Herrera, M., del, R. & Spooner, D. M. The single Andigenum origin of Neo-Tuberosum potato materials is not supported by microsatellite and plastid marker analyses. Theor. Appl. Genet. 118, 963–969 (2009).
The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011).
Spooner, D. M., Ghislain, M., Simon, R., Jansky, S. H. & Gavrilenko, T. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev. 80, 283–383 (2014).
Hamilton, J. P. et al. Single nucleotide polymorphism discovery in elite North American potato germplasm. BMC Genom. 12, 302 (2011).
Hirsch, C. N. et al. Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 3, 1003–1013 (2013).
Felcher, K. J. et al. Integration of two diploid potato linkage maps with the potato genome sequence. PLoS ONE 7, e36347 (2012).
Shan, J. et al. Transcriptome analysis reveals novel genes potentially involved in photoperiodic tuberization in potato. Genomics 102, 388–396 (2013).
Uitdewilligen, J. G. A. M. L. et al. A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS ONE 8, e62355 (2013).
Briggs, A. W. et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14616–14621 (2007).
Briggs, A. W. et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010).
Bukasov, S. M. Principles of the systematics of potatoes. Trudy po prikladnoi botanike, genetike i selektsii 62, 3–35 (1978).
Weiß, C. L., Pais, M., Cano, L. M., Kamoun, S. & Burbano, H. A. nQuire: a statistical framework for ploidy estimation using next generation sequencing. BMC Bioinformatics 19, 122 (2018).
Salaman, R. N. The early European potato: its character and place of origin. Bot. J. Linn. Soc. 53, 1–27 (1946).
Austin Bourke, P. M. Emergence of potato blight, 1843–46. Nature 203, 805 (1964).
Yoshida, K. et al. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. Elife 2, e00731 (2013).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Bradshaw, J. E., Bryan, G. J. & Ramsay, G. Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Res. 49, 49–65 (2006).
Hardigan, M. A. et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl Acad. Sci. USA 114, E9999–E10008 (2017).
Sawler, J. et al. Genomics assisted ancestry deconvolution in grape. PLoS ONE 8, e80791 (2013).
Bryc, K. et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl Acad. Sci. USA 107, 786–791 (2010).
Tajima, F. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Gao, H. et al. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genomics 19, 726 (2018).
Martínez-García, J. F., García-Martínez, J. L., Bou, J. & Prat, S. The interaction of gibberellins and photoperiod in the control of potato tuberization. J. Plant Growth Regul. 20, 377–386 (2001).
Walia, H. et al. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19, 1128–1132 (2009).
Köhler, C., Mittelsten Scheid, O. & Erilova, A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 26, 142–148 (2010).
Nielsen, R. et al. Tracing the peopling of the world through genomics. Nature 541, 302 (2017).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Choi, J. Y. et al. The rice paradox: multiple origins but single domestication in asian rice. Mol. Biol. Evol. 34, 11 (2017).
van Heerwaarden, J. et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl Acad. Sci. USA 108, 1088–92 (2011).
da Fonseca, R. R. et al. The origin and evolution of maize in the Southwestern United States. Nat. Plants 1, 14003 (2015).
Gutaker, R. M., Reiter, E., Furtwängler, A., Schuenemann, V. J. & Burbano, H. A. Extraction of ultrashort DNA molecules from herbarium specimens. Biotechniques 62, 76–79 (2017).
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, db.prot5448 (2010).
Meyer, M. et al. A high-coverage genome sequence from an archaic denisovan individual. Science 338, 222–226 (2012).
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012).
Hodges, E. et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 4, 960–974 (2009).
Burbano, H. A. et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725 (2010).
Jiang, H., Lei, R., Ding, S.-W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 1–12 (2014).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Hofreiter, M., Jaenicke, V., Serre, D., von Haeseler, A. & Pääbo, S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 29, 4793–4799 (2001).
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013).
R Core Team A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014).
Peltzer, A. et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016).
100 Tomato Genome Sequencing Consortium. et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80, 136–148 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Scrucca, L., Fop, M., Murphy, T. B. & Raftery, A. E. mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J. 8, 289–317 (2016).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–33 (2013).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Mardia, K. V. Some properties of clasical multi-dimesional scaling. Commun. Stat. Theory Methods 7, 1233–1241 (1978).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Leppälä, K., Nielsen, S. V. & Mailund, T. admixturegraph: an R package for admixture graph manipulation and fitting. Bioinformatics 33, 1738–1740 (2017).
Paradis, E. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420 (2010).
Acknowledgements
We thank D. Weigel for initial discussions during the conception of this project; B. Glover and C. Bartram (Cambridge University Herbarium), M. Graniszewska and H. Leśniewska (University of Warsaw) for granting access to historic herbarium samples; J. Krause (MPI for the Science of Human History), V. Schuenemann and E. Reiter (University of Tuebingen) for access to clean-room facilities and technical support; M. Neumann and S. Latorre for laboratory assistance; L. Shannon (University of Minnesota) for input on potato diversity; C. Bachem, H. van Eck and J. Willemsen (Wageningen University) for input on array design; N. Arciniegas (National University of Colombia) for collection assistance; T. Karasov, M. Zaidem and members of the Burbano laboratory for comments on the manuscript; and M. Purugganan (New York University) for supporting R. Gutaker during the final stages of the project. We thank the Spanish National Research Council (CSIC) and the Ministry of Economy and Competitiveness for financing the project No. CGL 2010–19747, which facilitated the visits to Colombia and access to the collections of the Real Jardin Botanico Herbarium (MA). This study was funded by the Max Planck Society and its Presidential Innovation Fund (H.A.B).
Author information
Authors and Affiliations
Contributions
R.M.G and H.A.B. conceived and designed the study with input from S.P. D.E., N.L.A., S.K., J.L.F.-A. and S.P curated, identified, pre-selected and contributed contemporary and historic potato samples. R.M.G carried out DNA extractions, hybridization captures and library preparations. R.M.G. performed the bioinformatic processing of sequencing data with input from C.L.W and H.A.B. C.L.W. carried out ploidy estimation of samples. R.M.G. performed the phylogeographic and population genomics analyses with input from H.A.B. R.M.G., S.P. and H.A.B. contributed to the interpretation of the data. R.M.G and H.A.B wrote the manuscript with input from all authors. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Figs. 1–15
Supplementary Tables 1
Provenance and meta-information of historical samples.
Supplementary Tables 2
Provenance and meta-information of modern samples.
Supplementary Tables 3
Sequenced loci nuclear.
Supplementary Tables 4
Characteristics of sequenced libraries.
Supplementary Tables 5
Read support for different StCDF1 alleles.
Supplementary Tables 6
Loss of heterozygosity and Tajima’s D-statistics.
Supplementary Tables 7
Description: evaluation of the 100 best admixture graph models.
Rights and permissions
About this article
Cite this article
Gutaker, R.M., Weiß, C.L., Ellis, D. et al. The origins and adaptation of European potatoes reconstructed from historical genomes. Nat Ecol Evol 3, 1093–1101 (2019). https://doi.org/10.1038/s41559-019-0921-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0921-3
This article is cited by
-
Diversity and population structure of Nordic potato cultivars and breeding clones
BMC Plant Biology (2022)
-
Resequencing and genome-wide association studies of autotetraploid potato
Molecular Horticulture (2022)
-
The genetic architectures of vine and skin maturity in tetraploid potato
Theoretical and Applied Genetics (2022)
-
Diploid Potatoes as a Catalyst for Change in the Potato Industry
American Journal of Potato Research (2022)
-
DNA methylation affects photoperiodic tuberization in potato (Solanum tuberosum L.) by mediating the expression of genes related to the photoperiod and GA pathways
Horticulture Research (2021)