Abstract
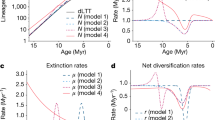
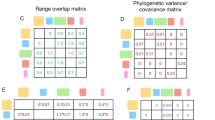
Understanding how and why diversification rates vary through time and space and across species groups is key to understanding the emergence of today’s biodiversity. Phylogenetic approaches aimed at identifying variations in diversification rates during the evolutionary history of clades have focused on exceptional shifts subtending evolutionary radiations. While such shifts have undoubtedly affected the history of life, identifying smaller but more frequent changes is important as well. We developed ClaDS—a new Bayesian approach for estimating branch-specific diversification rates on a phylogeny that relies on a model with changes in diversification rates at each speciation event. We show, using Monte Carlo simulations, that the approach performs well at inferring both small and large changes in diversification. Applying our approach to bird phylogenies covering the entire avian radiation, we find that diversification rates are remarkably heterogeneous within evolutionarily restricted species groups. Some groups such as Accipitridae (hawks and allies) cover almost the full range of speciation rates found across the entire bird radiation. As much as 76% of the variation in branch-specific rates across this radiation is due to intraclade variation, suggesting that a large part of the variation in diversification rates is due to many small, rather than few large, shifts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The simulated phylogenies used to test the method are available at https://github.com/OdileMaliet/ClaDS/tree/master/Simulations in a file named trees.zip. All of the empirical data used for the analysis were obtained from the Jetz et al.4 study, and are available from https://www.nature.com/articles/nature11631.
Code availability
The R functions used to simulate and fit the model are available in the RPANDA R package. All of the codes used to test our method are available from the GitHub repository at https://github.com/OdileMaliet/ClaDS.git.
References
Alfaro, M. E. et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13410–13414 (2009).
Chan, K. M. & Moore, B. R. SymmeTREE: whole-tree analysis of differential diversification rates. Bioinformatics 21, 1709–1710 (2004).
Morlon, H., Parsons, T. L. & Plotkin, J. B. Reconciling molecular phylogenies with the fossil record. Proc. Natl Acad. Sci. USA 108, 16327–16332 (2011).
Jetz, W., Thomas, G., Joy, J., Hartmann, K. & Mooers, A. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Rabosky, D. L. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 (2014).
Barido-Sottani, J., Vaughan, T. G. & Stadler, T. A multi-state birth–death model for Bayesian inference of lineage-specific birth and death rates. Preprint at https://www.biorxiv.org/content/10.1101/440982v1 (2018).
Sanderson, M. J. & Wojciechowski, M. F. Diversification rates in a temperate legume clade: are there so many species of Astragalus (Fabaceae)? Am. J. Bot. 83, 1488–1502 (1996).
Miller, A. H. In Ornithologie als Biologische Wissenschaft (eds Mayr, E. and Schüz, E.) 84–88 (Carl Winter, 1949).
Hunter, J. P. Key innovations and the ecology of macroevolution. Trends Ecol. Evol. 13, 31–36 (1998).
Benton, M. J. The Red Queen and the court jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732 (2009).
Goldberg, E. E. et al. Species selection maintains self-incompatibility. Science 330, 493–495 (2010).
Onstein, R. E. et al. Frugivory-related traits promote speciation of tropical palms. Nat. Ecol. Evol. 1, 1903–1911 (2017).
FitzJohn, R. G. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092 (2012).
Title, P. O. & Rabosky, D. L. Tip rates, phylogenies, and diversification: what are we estimating, and how good are the estimates? Methods Ecol. Evol. https://doi.org/10.1111/2041-210X.13153 (2018).
Emerson, B. C. & Kolm, N. Species diversity can drive speciation. Nature 434, 1015–1017 (2005).
Rabosky, D. L. & Lovette, I. J. Explosive evolutionary radiations: decreasing speciation or increasing extinction through time? Evolution 62, 1866–1875 (2008).
Phillimore, A. B. & Price, T. D. Density-dependent cladogenesis in birds. PLoS Biol. 6, e71 (2008).
Moen, D. & Morlon, H. Why does diversification slow down? Trends Ecol. Evol. 29, 190–197 (2014).
Rosenzweig, M. L. Species diversity gradients: we know more and less than we thought. J. Mammal. 73, 715–730 (1992).
Rabosky, D. L. & Huang, H. A robust semi-parametric test for detecting trait-dependent diversification. Syst. Biol. 65, 181–193 (2015).
May, M. R. & Moore, B. R. How well can we detect lineage-specific diversification-rate shifts? A simulation study of sequential AIC methods. Syst. Biol. 65, 1076–1084 (2016).
Moore, B. R., Höhna, S., May, M. R., Rannala, B. & Huelsenbeck, J. P. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl Acad. Sci. USA 113, 9569–9574 (2016).
Rabosky, D. L., Mitchell, J. S. & Chang, J. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst. Biol. 66, 477–498 (2017).
Rabosky, D. L. How to make any method “fail”: BAMM at the kangaroo court of false equivalency. Preprint at https://arxiv.org/abs/1711.03253 (2017).
Rabosky, D. L. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824 (2010).
Mitchell, J. S. & Rabosky, D. L. Bayesian model selection with BAMM: effects of the model prior on the inferred number of diversification shifts. Methods Ecol. Evol. 8, 37–46 (2017).
Freckleton, R. P., Phillimore, A. B. & Pagel, M. Relating traits to diversification: a simple test. Am. Nat. 172, 102–115 (2008).
Rabosky, D. L. & Goldberg, E. E. FiSSE: a simple nonparametric test for the effects of a binary character on lineage diversification rates. Evolution 71, 1432–1442 (2017).
Harvey, M. G. & Rabosky, D. L. Continuous traits and speciation rates: alternatives to state-dependent diversification models. Methods Ecol. Evol. 9, 984–993 (2017).
Bromham, L., Hua, X. & Cardillo, M. Detecting macroevolutionary self-destruction from phylogenies. Syst. Biol. 65, 109–127 (2015).
Hua, X. & Bromham, L. Phylometrics: an R package for detecting macroevolutionary patterns, using phylogenetic metrics and backward tree simulation. Methods Ecol. Evol. 7, 806–810 (2016).
Maddison, W. P., Midford, P. E. & Otto, S. P. Estimating a binary character’s effect on speciation and extinction. Syst. Biol. 56, 701–710 (2007).
Redding, D. W. & Mooers, A. Ø. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678 (2006).
Beaulieu, J. M. & O’Meara, B. C. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601 (2016).
Nee, S., May, R. M. & Harvey, P. H. The reconstructed evolutionary process. Phil. Trans. R. Soc. Lond. B 344, 305–311 (1994).
Stadler, T. Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl Acad. Sci. USA 108, 6187–6192 (2011).
Condamine, F. L., Rolland, J. & Morlon, H. Macroevolutionary perspectives to environmental change. Ecol. Lett. 16, 72–85 (2013).
Lewitus, E. & Morlon, H. Detecting environment-dependent diversification from phylogenies: a simulation study and some empirical illustrations. Syst. Biol. 67, 576–593 (2017).
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015).
Thorne, J. L., Kishino, H. & Painter, I. S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 15, 1647–1657 (1998).
Huelsenbeck, J. P., Larget, B. & Swofford, D. A compound Poisson process for relaxing the molecular clock. Genetics 154, 1879–1892 (2000).
Lartillot, N., Phillips, M. J. & Ronquist, F. A mixed relaxed clock model. Phil. Trans. R. Soc. B 371, 20150132 (2016).
Morlon, H. et al. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol. 7, 589–597 (2016).
Yang, Z. & Rodríguez, C. E. Searching for efficient Markov chain Monte Carlo proposal kernels. Proc. Natl Acad. Sci. USA 110, 19307–19312 (2013).
Gelman, A et al. Bayesian Data Analysis (CRC press, 2014).
Ter Braak, C. J. & Vrugt, J. A. Differential evolution Markov chain with snooker updater and fewer chains. Stat. Comput. 18, 435–446 (2008).
Grafen, A. The phylogenetic regression. Phil. Trans. R. Soc. Lond. B 326, 119–157 (1989).
Acknowledgements
The authors are very grateful to L. Arístide, J. Clavel, J. Drury, C. Fruciano, S. Lambert, E. Lewitus, M. Manceau, O. Missa, B. Perez, A. Catarina Silva and G. Sommeria-Klein for their helpful comments on an earlier version of this manuscript. This work was supported by an AMX grant (from École Polytechique) and the LabEx MemoLife (to O.M.), PROCOPE Mobility Grant 57134817 (to F.H. and H.M.) and the European Research Council (ERC 616419-PANDA to H.M.).
Author information
Authors and Affiliations
Contributions
O.M., F.H. and H.M. designed the study and performed research. O.M. contributed new analytical tools and analysed the data. O.M., F.H. and H.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Results and Figs. 1–34
Rights and permissions
About this article
Cite this article
Maliet, O., Hartig, F. & Morlon, H. A model with many small shifts for estimating species-specific diversification rates. Nat Ecol Evol 3, 1086–1092 (2019). https://doi.org/10.1038/s41559-019-0908-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0908-0
This article is cited by
-
Analysis of intrinsic evolutionary factors leading to microendemic distributions in New Caledonian leaf beetles
Scientific Reports (2023)
-
The importance of the Andes in the evolutionary radiation of Sigmodontinae (Rodentia, Cricetidae), the most diverse group of mammals in the Neotropics
Scientific Reports (2023)
-
Investigating the reliability of molecular estimates of evolutionary time when substitution rates and speciation rates vary
BMC Ecology and Evolution (2022)
-
Phylogenomic analysis of Syngnathidae reveals novel relationships, origins of endemic diversity and variable diversification rates
BMC Biology (2022)
-
Universal probabilistic programming offers a powerful approach to statistical phylogenetics
Communications Biology (2021)