Abstract
Admixture is a recurrent phenomenon in humans and other great ape populations. Genetic information from extinct hominins allows us to study historical interactions with modern humans and discover adaptive functions of gene flow. Here, we investigate whole genomes from bonobo and chimpanzee populations for signatures of gene flow from unknown archaic populations, finding evidence for an ancient admixture event between bonobos and a divergent lineage. This result reveals a complex population history in our closest living relatives, probably several hundred thousand years ago. We reconstruct up to 4.8% of the genome of this ‘ghost’ ape, which represents genomic data of an extinct great ape population. Genes contained in archaic fragments might confer functional consequences for the immunity, behaviour and physiology of bonobos. Finally, comparing the landscapes of introgressed regions in humans and bonobos, we find that a recurrent depletion of introgression is rare, suggesting that genomic incompatibilities arose seldom in these lineages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Sequence data from a previous study are publicly available under the accession code PRJEB15086 at the European Nucleotide Archive. Genotype data are available at http://biologiaevolutiva.org/tmarques/data/. Data pertaining to the results are in the Supplementary Information.
Change history
14 May 2019
In the version of this article originally published, a funding acknowledgement was missing for Tomas Maques-Bonet. The original funding statement was: “T.M.-B. was supported by MINECO BFU2014-55090-P (FEDER), a U01 MH106874 grant, the Howard Hughes International Early Career programme, Obra Social ‘La Caixa’ and Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya.” It has been updated to: “T.M.-B. was supported by BFU2017-86471-P (MINECO/FEDER, UE), a U01 MH106874 grant, the Howard Hughes International Early Career programme, Obra Social ‘La Caixa’ and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880).” The error has been corrected in the HTML and PDF versions of this article.
References
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010).
Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010).
Hammer, M. F., Woerner, A. E., Mendez, F. L., Watkins, J. C. & Wall, J. D. Genetic evidence for archaic admixture in Africa. Proc. Natl Acad. Sci. USA 108, 15123–15128 (2011).
Meyer, M. et al. A high coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012).
Vernot, B. & Akey, J. M. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021 (2014).
Fu, Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015).
Vernot, B. et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235–239 (2016).
Xu, D. et al. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol. Biol. Evol. 34, 2704–2715 (2017).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Kuhlwilm, M. et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429–433 (2016).
Posth, C. et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat Commun. 8, 16046 (2017).
Juric, I., Aeschbacher, S. & Coop, G. The strength of selection against Neanderthal introgression. PLoS Genet. 12, e1006340 (2016).
Sankararaman, S., Mallick, S., Patterson, N. & Reich, D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26, 1241–1247 (2016).
Huerta-Sanchez, E. et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014).
Racimo, F., Marnetto, D. & Huerta-Sánchez, E. Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol. 34, 296–317 (2017).
Brunet, M. et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature 418, 145–151 (2002).
McBrearty, S. & Jablonski, N. G. First fossil chimpanzee. Nature 437, 105–108 (2005).
Prado-Martinez, J. et al. Great ape genetic diversity and population history. Nature 499, 471–475 (2013).
Hey, J. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol. Biol. Evol. 27, 921–933 (2010).
Tung, J. & Barreiro, L. B. The contribution of admixture to primate evolution. Curr. Opin. Genet. Dev. 47, 61–68 (2017).
De Manuel, M. et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354, 477–481 (2016).
Kawamoto, Y. et al. Genetic structure of wild bonobo populations: diversity of mitochondrial DNA and geographical distribution. PLoS One 8, e59660 (2013).
Takemoto, H., Kawamoto, Y. & Furuichi, T. How did bonobos come to range south of the Congo river? Reconsideration of the divergence of Pan paniscus from other Pan populations. Evol. Anthropol. 24, 170–184 (2015).
Takemoto, H. et al. The mitochondrial ancestor of bonobos and the origin of their major haplogroups. PLoS One 12, e0174851 (2017).
Myers Thompson, J. A. A model of the biogeographical journey from proto-Pan to Pan paniscus. Primates 44, 191–197 (2003).
Skov, L. et al. Detecting archaic introgression using an unadmixed outgroup. PLoS Genet. 14, e1007641 (2018).
Won, Y.-J. J. & Hey, J. Divergence population genetics of chimpanzees. Mol. Biol. Evol. 22, 297–307 (2005).
Plagnol, V. & Wall, J. D. Possible ancestral structure in human populations. PLoS Genet. 2, e105 (2006).
Wall, J. D., Lohmueller, K. E. & Plagnol, V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol. Biol. Evol. 26, 1823–1827 (2009).
Hsieh, P. et al. Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies. Genome Res. 26, 291–300 (2016).
Lachance, J. et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell 150, 457–469 (2012).
Nye, J. et al. Selection in the introgressed regions of the chimpanzee genome. Genome Biol. Evol. 10, 1132–1138 (2018).
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., Sousa, V. C. & Foll, M. Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 (2013).
Hey, J. et al. Phylogeny estimation by integration over isolation with migration models. Mol. Biol. Evol. 35, 2805–2818 (2018).
Prüfer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017).
Besenbacher, S., Hvilsom, C., Marques-Bonet, T., Mailund, T. & Schierup, M. H. Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat. Ecol. Evol. 3, 286–292 (2019).
Rasmussen, M. D., Hubisz, M. J., Gronau, I. & Siepel, A. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 10, e1004342 (2014).
Prüfer, K. et al. The bonobo genome compared with the chimpanzee and human genomes. Nature 486, 527–531 (2012).
Kuhlwilm, M. et al. Evolution and demography of the great apes. Curr. Opin. Genet. Dev. 41, 124–129 (2016).
Sankararaman, S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357 (2014).
Nam, K. et al. Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc. Natl Acad. Sci. USA 112, 6413–6418 (2015).
Steinrücken, M., Spence, J. P., Kamm, J. A., Wieczorek, E. & Song, Y. S. Model-based detection and analysis of introgressed Neanderthal ancestry in modern humans. Mol. Ecol. 27, 3873–3888 (2018).
Piccinni, M.-P. T cells in normal pregnancy and recurrent pregnancy loss. Reprod. Biomed. Online 13, 840–844 (2006).
Han, S., Andrés, A. M., Marques-Bonet, T. & Kuhlwilm, M. Genetic variation in Pan species is shaped by demographic history and harbors lineage-specific functions. Genome Biol. Evol. https://doi.org/10.1093/gbe/evz047 (2019).
Kuhlwilm, M. & Boeckx, C. Genetic differences between humans and other hominins contribute to the “human condition”. Preprint at https://www.biorxiv.org/content/10.1101/298950v1 (2018).
Furuichi, T. Social interactions and the life history of female Pan paniscus in Wamba, Zaire. Int. J. Primatol. 10, 173–197 (1989).
Cagan, A. et al. Natural selection in the great apes. Mol. Biol. Evol. 33, 3268–3283 (2016).
Brawand, D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011).
Bouhassira, E. E. et al. An alanine-to-threonine substitution in protein 4.2 cDNA is associated with a Japanese form of hereditary hemolytic anemia (protein 4.2NIPPON). Blood 79, 1846–1854 (1992).
Loomis, M. R. in Zoo and Wild Animal Medicine 5th edn (eds Fowler, M. E. & Miller, R. E.) 381–397 (Saunders (Elsevier Science), 2003).
Cyrklaff, M. et al. Hemoglobins S and C interfere with actin remodeling in Plasmodium falciparum-infected erythrocytes. Science 334, 1283–1286 (2011).
Dannemann, M., Andrés, A. M. & Kelso, J. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am. J. Hum. Genet. 98, 22–33 (2016).
Frazer, J. K. et al. Identification of centerin: a novel human germinal center B cell-restricted serpin. Eur. J. Immunol. 30, 3039–3048 (2000).
Suzuki, K. et al. A novel glycosylphosphatidyl inositol-anchored protein on human leukocytes: a possible role for regulation of neutrophil adherence and migration. J. Immunol. 162, 4277–4284 (1999).
Hernandez-Rodriguez, J. et al. The impact of endogenous content, replicates and pooling on genome capture from faecal samples. Mol. Ecol. Resour. 18, 319–333 (2017).
Paten, B. et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 18, 1829–1843 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Lawrence, M., Gentleman, R. & Carey, V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009).
R Core Development Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015).
Vernot, B. freezing-archer (2016), GitHub repository, https://github.com/bvernot/freezing-archer.
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Haider, S. et al. A bedr way of genomic interval processing. Source Code Biol. Med. 11, 14 (2016).
Wood, S. N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 73, 3–36 (2011).
Hudson, R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012).
Langergraber, K. E. et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl Acad. Sci. USA 109, 15716–15721 (2012).
Csilléry, K., François, O. & Blum, M. G. B. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3, 475–479 (2012).
Stevison, L. S. et al. The time-scale of recombination rate evolution in great apes. Mol. Biol. Evol. 33, 928–945 (2016).
Kelleher, J., Etheridge, A. M. & McVean, G. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Comput. Biol. 12, e1004842 (2016).
Schliep, K. P. phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011).
Jombart, T. & Ahmed, I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Paradis, E. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420 (2010).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Pfeifer, B., Wittelsbürger, U., Ramos-Onsins, S. E. & Lercher, M. J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936 (2014).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Acknowledgements
We thank A. M. Andres, B. Vernot and L. J. Kuhlwilm for comments and discussion, and M. de Manuel for help with the data. M.K. was supported by a DFG fellowship (KU 3467/1-1). V.C.S. was supported by the Fundação para a Ciência e a Tecnologia (project UID/BIA/00329/2013) and EU Horizon 2020 programme (Marie Skłodowska-Curie grant 799729). L.E. was supported by the Swiss National Science Foundation (number 310030B-166605). T.M.-B. was supported by BFU2017-86471-P (MINECO/FEDER, UE), a U01 MH106874 grant, the Howard Hughes International Early Career programme, Obra Social "La Caixa" and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880).
Author information
Authors and Affiliations
Contributions
M.K., S.H., V.C.S. and L.E. analysed the data. M.K. and T.M.-B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods and analysis, Supplementary Figures 1–32
Supplementary Tables
Supplementary Tables 1–21
Supplementary Data
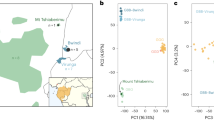
Principal component analysis (PCA) of putatively introgressed fragments.
Rights and permissions
About this article
Cite this article
Kuhlwilm, M., Han, S., Sousa, V.C. et al. Ancient admixture from an extinct ape lineage into bonobos. Nat Ecol Evol 3, 957–965 (2019). https://doi.org/10.1038/s41559-019-0881-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0881-7
This article is cited by
-
Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution
Nature Reviews Genetics (2023)
-
Ghost admixture in eastern gorillas
Nature Ecology & Evolution (2023)
-
Merging morphological and genetic evidence to assess hybridization in Western Eurasian late Pleistocene hominins
Nature Ecology & Evolution (2022)
-
Genomic variation from an extinct species is retained in the extant radiation following speciation reversal
Nature Ecology & Evolution (2022)