Abstract
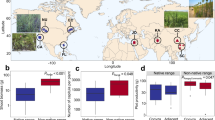
Plant range expansion is occurring at a rapid pace, largely in response to human-induced climate warming. Although the movement of plants along latitudinal and altitudinal gradients is well-documented, effects on belowground microbial communities remain largely unknown. Furthermore, for range expansion, not all plant species are equal: in a new range, the relatedness between range-expanding plant species and native flora can influence plant–microorganism interactions. Here we use a latitudinal gradient spanning 3,000 km across Europe to examine bacterial and fungal communities in the rhizosphere and surrounding soils of range-expanding plant species. We selected range-expanding plants with and without congeneric native species in the new range and, as a control, the congeneric native species, totalling 382 plant individuals collected across Europe. In general, the status of a plant as a range-expanding plant was a weak predictor of the composition of bacterial and fungal communities. However, microbial communities of range-expanding plant species became more similar to each other further from their original range. Range-expanding plants that were unrelated to the native community also experienced a decrease in the ratio of plant pathogens to symbionts, giving weak support to the enemy release hypothesis. Even at a continental scale, the effects of plant range expansion on the belowground microbiome are detectable, although changes to specific taxa remain difficult to decipher.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information. Sequences have been deposited in the European Nucleotide Archive under accession numbers PRJEB25697, PRJEB25694, PRJEB25693 and PRJEB25692.
References
Pecl, G. T. et al. Biodiversity redistribution under climate change: impacts on ecosystems and human well-being. Science 355, eaai9214 (2017).
Parmesan, C. & Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 (2003).
Classen, A. T. et al. Direct and indirect effects of climate change on soil microbial and soil microbial–plant interactions: what lies ahead? Ecosphere 6, art130 (2015).
Meisner, A., De Deyn, G. B., de Boer, W. & van der Putten, W. H. Soil biotic legacy effects of extreme weather events influence plant invasiveness. Proc. Natl Acad. Sci. USA 110, 9835–9838 (2013).
Engelkes, T. et al. Successful range-expanding plants experience less above-ground and below-ground enemy impact. Nature 456, 946–948 (2008).
van der Putten, W. H., Bradford, M. A., Brinkman, E. P., van de Voorde, T. F. J. & Veen, G. F. Where, when and how plant–soil feedback matters in a changing world. Funct. Ecol. 30, 1109–1121 (2016).
Gonzalez-Megias, A. & Menendez, R. Climate change effects on above- and below-ground interactions in a dryland ecosystem. Phil. Trans. R. Soc. B 367, 3115–3124 (2012).
Tylianakis, J. M., Didham, R. K., Bascompte, J. & Wardle, D. A. Global change and species interactions in terrestrial ecosystems. Ecol. Lett. 11, 1351–1363 (2008).
Kourtev, P. S., Ehrenfeld, J. G. & Häggblom, M. Exotic plant species alter the microbial community structure and function in the soil. Ecology 83, 3152–3166 (2002).
McLeod, M. L. et al. Exotic invasive plants increase productivity, abundance of ammonia-oxidizing bacteria and nitrogen availability in intermountain grasslands. J. Ecol. 104, 994–1002 (2016).
Coats, V. C. & Rumpho, M. E. The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front. Microbiol. 5, 368 (2014).
Klironomos, J. N. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70 (2002).
Kardol, P. & Wardle, D. A. How understanding aboveground–belowground linkages can assist restoration ecology. Trends Ecol. Evol. 25, 670–679 (2010).
Van Nuland, M. E., Bailey, J. K. & Schweitzer, J. A. Divergent plant–soil feedbacks could alter future elevation ranges and ecosystem dynamics. Nat. Ecol. Evol. 1, 0150 (2017).
van Grunsven, R. H. A. et al. Reduced plant–soil feedback of plant species expanding their range as compared to natives. J. Ecol. 95, 1050–1057 (2007).
Dostálek, T., Münzbergová, Z., Kladivová, A. & Macel, M. Plant–soil feedback in native vs. invasive populations of a range expanding plant. Plant Soil 399, 209–220 (2015).
Berg, G. & Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13 (2009).
De Frenne, P. et al. Plant movements and climate warming: intraspecific variation in growth responses to nonlocal soils. New Phytol. 202, 431–441 (2014).
Van Grunsven, R. H. A., van der Putten, W. H., Bezemer, T., Berendse, F. & Veenendaal, E. M. Plant–soil interactions in the expansion and native range of a poleward shifting plant species. Glob. Change Biol. 16, 380–385 (2010).
Collins, C. G., Carey, C. J., Aronson, E. L., Kopp, C. W. & Diez, J. M. Direct and indirect effects of native range expansion on soil microbial community structure and function. J. Ecol. 104, 1271–1283 (2016).
Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017).
Kuramae, E., Gamper, H., van Veen, J. & Kowalchuk, G. Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiol. Ecol. 77, 285–294 (2011).
Tecon, R. & Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 41, 599–623 (2017).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Delgado-Baquerizo, M. et al. A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018).
Talbot, J. M. et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl Acad. Sci. USA 111, 6341–6346 (2014).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Barberán, A. et al. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802 (2014).
Lekberg, Y., Rosendahl, S. & Olsson, P. A. The fungal perspective of arbuscular mycorrhizal colonization in ‘nonmycorrhizal’ plants. New Phytol. 205, 1399–1403 (2015).
Lau, J. A. & Lennon, J. T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl Acad. Sci. USA 109, 14058–14062 (2012).
de Vries, F. T. et al. Land use alters the resistance and resilience of soil food webs to drought. Nat. Clim. Change 2, 276–280 2012).
Peay, K. G. Back to the future: natural history and the way forward in modern fungal ecology. Fungal Ecol. 12, 4–9 (2014).
Edwards, J. et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl Acad. Sci. USA 112, E911–E920 (2015).
Pieterse, C. M. J., de Jonge, R. & Berendsen, R. L. The soil-borne supremacy. Trends Plant Sci. 21, 171–173 (2016).
Prober, S. M. et al. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95 (2015).
Gilbert, G. S. & Webb, C. O. Phylogenetic signal in plant pathogen–host range. Proc. Natl Acad. Sci. USA 104, 4979–4983 (2007).
Parker, I. M. et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542–544 (2015).
Lankau, R. A. Coevolution between invasive and native plants driven by chemical competition and soil biota. Proc. Natl Acad. Sci. USA 109, 11240–11245 (2012).
Morriën, E. et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 8, 14349 (2017).
Keymer, D. P. & Lankau, R. A. Disruption of plant–soil–microbial relationships influences plant growth. J. Ecol. 105, 816–827 (2017).
Leach, J. E., Triplett, L. R., Argueso, C. T. & Trivedi, P. Communication in the phytobiome. Cell 169, 587–596 (2017).
Bakkenes, M., Alkemade, J. R. M., Ihle, F., Leemans, R. & Latour, J. B. Assessing effects of forecasted climate change on the diversity and distribution of European higher plants for 2050. Glob. Change Biol. 8, 390–407 (2002).
Wilschut, R. A., Kostenko, O., Koorem, K. & van der Putten, W. H. Nematode community responses to range-expanding and native plant communities in original and new range soils. Ecol. Evol. 8, 10288–10297 (2018).
Koorem, K. et al. Relatedness with plant species in native community influences ecological consequences of range expansions. Oikos 127, 981–990 (2018).
van der Heijden, M. G. A. & Hartmann, M. Networking in the plant microbiome. PLoS Biol. 14, e1002378 (2016).
Leff, J. W. et al. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 12, 1794–1805 (2018).
Fierer, N. et al. Reconstructing the Microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342, 621–624 (2013).
Emmett, B. D., Youngblut, N. D., Buckley, D. H. & Drinkwater, L. E. Plant phylogeny and life history shape rhizosphere bacterial microbiome of summer annuals in an agricultural field. Front. Microbiol. 8, 2414 (2017).
Goberna, M., Navarro-Cano, J. A. & Verdú, M. Opposing phylogenetic diversity gradients of plant and soil bacterial communities. Proc. R. Soc. B 283, 20153003 (2016).
Reynolds, H. L., Packer, A., Bever, J. D. & Clay, K. Grassroots ecology: plant–microbe–soil interactions as drivers of plant community structure and dynamics. Ecology 84, 2281–2291 (2003).
Bennett, J. A. et al. Plant–soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184 (2017).
Golivets, M. & Wallin, K. F. Neighbour tolerance, not suppression, provides competitive advantage to non-native plants. Ecol. Lett. 21, 745–759 (2018).
Geml, J. & Wagner, M. R. Out of sight, but no longer out of mind — towards an increased recognition of the role of soil microbes in plant speciation. New Phytol. 217, 965–967 (2018).
Dawson, W. Release from belowground enemies and shifts in root traits as interrelated drivers of alien plant invasion success: a hypothesis. Ecol. Evol. 5, 4505–4516 (2015).
Blumenthal, D., Mitchell, C. E., Pysek, P. & Jarosík, V. Synergy between pathogen release and resource availability in plant invasion. Proc. Natl Acad. Sci. USA 106, 7899–7904 (2009).
Wilschut, R. A., Silva, J. C. P., Garbeva, P. & van der Putten, W. H. Belowground plant–herbivore interactions vary among climate-driven range-expanding plant species with different degrees of novel chemistry. Front. Plant Sci. 8, 1861 (2017).
Inderjit & van der Putten, W. H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 25, 512–519 (2010).
Rout, M. E. & Callaway, R. M. Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that ‘everything is not everywhere’. Ann. Bot 110, 213–222 (2012).
Nguyen, N. H. et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016).
van der Putten, W. H. Climate change, aboveground–belowground interactions, and species’ range shifts. Annu. Rev. Ecol. Evol. Syst. 43, 365–383 (2012).
Mitchell, C. E. & Power, A. G. Release of invasive plants from fungal and viral pathogens. Nature 421, 625–627 (2003).
Bever, J. D., Mangan, S. A. & Alexander, H. M. Maintenance of plant species diversity by pathogens. Annu. Rev. Ecol. Evol. Syst. 46, 305–325 (2015).
Morriën, E. et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 8, 14349 (2017).
Hannula, S. E. et al. Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J. 11, 2294–2304 (2017).
Jousset, A. et al. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862 (2017).
van Kleunen, M., Dawson, W. & Maurel, N. Characteristics of successful alien plants. Mol. Ecol. 24, 1954–1968 (2015).
Chen, I.-C., Hill, J. K., Ohlemuller, R., Roy, D. B. & Thomas, C. D. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011).
Bever, J., Platt, T. & Morton, E. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 66, 265–283 (2012).
Wubs, E. R. J., van der Putten, W. H., Bosch, M. & Bezemer, T. M. Soil inoculation steers restoration of terrestrial ecosystems. Nat. Plants 2, 16107 (2016).
Alexander, J. M., Diez, J. M. & Levine, J. M. Novel competitors shape species’ responses to climate change. Nature 525, 515–518 (2015).
Fordham, D. A. et al. Plant extinction risk under climate change: are forecast range shifts alone a good indicator of species vulnerability to global warming? Glob. Change Biol. 18, 1357–1371 (2012).
Tamis, W. L. M., van’t Zelfde, M., van der Meijden, R. & de Haes, H. A. U. Changes in vascular plant biodiversity in the Netherlands in the 20th century explained by their climatic and other environmental characteristics. Climatic Change 72, 37–56 (2005).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Ihrmark, K. et al. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677 (2012).
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D. & Dangl, J. L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002 (2013).
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G. & Neufeld, J. D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13, 31 (2012).
Dodt, M., Roehr, J., Ahmed, R. & Dieterich, C. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1, 895–905 (2012).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
Bengtsson-Palme, J. et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919 (2013).
Kõljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013).
Koster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.0-10 https://cran.r-project.org/web/packages/vegan/index.html (2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Acknowledgements
We are grateful for the support of Ž. Modrić-Surina, S. Dragićević, I. Starke and M. Hohla, who all helped with sampling. This work was supported in large part by the European Research Council (ERC advanced grant ERC-Adv 323020 (SPECIALS) to W.H.v.d.P. Additional support came from the Estonian Research Council (grant PUTJD78) (K.K.) and the Slovenian Research Agency (research core funding no. P1-0236) (B.V. and T.Č.).
Author information
Authors and Affiliations
Contributions
W.H.v.d.P. conceived the idea of this study. Sample collection was completed W.H.v.d.P., K.S.R., K.K., S.G., L.J.B., F.t.H., O.K., N.K., M.M., D.C., M.A.T., B.V., T.Č., C.W. and R.A.W. Soil analyses and sequencing were completed by L.J.B., F.t.H., C.W., D.v.R. and K.S.R. Data analyses were completed by L.B.S. and K.S.R. The manuscript was written by K.S.R., with contributions from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–7 and Supplementary Figures 1–3
Supplementary Data 1
OTU tables: microbial taxa (OTUs) abundances
Supplementary Data 2
Environmental Factors by sample: all samples with plant information, soil abiotic factors and locations
Rights and permissions
About this article
Cite this article
Ramirez, K.S., Snoek, L.B., Koorem, K. et al. Range-expansion effects on the belowground plant microbiome. Nat Ecol Evol 3, 604–611 (2019). https://doi.org/10.1038/s41559-019-0828-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0828-z
This article is cited by
-
Microbiome specificity and fluxes between two distant plant taxa in Iberian forests
Environmental Microbiome (2023)
-
Taxonomic and environmental distribution of bacterial amino acid auxotrophies
Nature Communications (2023)
-
Species composition of root-associated mycobiome of ruderal invasive Anthemis cotula L. varies with elevation in Kashmir Himalaya
International Microbiology (2023)
-
Legacy effects post removal of a range-expanding shrub influence soil fungal communities and create negative plant-soil feedbacks for conspecific seedlings
Plant and Soil (2023)
-
Plant-microbe interactions in the phyllosphere: facing challenges of the anthropocene
The ISME Journal (2022)