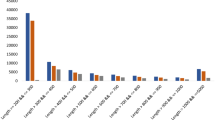
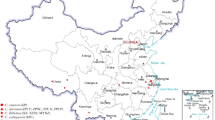
Abstract
New protein-coding genes that arise de novo from non-coding DNA sequences contribute to protein diversity. However, de novo gene origination is challenging to study as it requires high-quality reference genomes for closely related species, evidence for ancestral non-coding sequences, and transcription and translation of the new genes. High-quality genomes of 13 closely related Oryza species provide unprecedented opportunities to understand de novo origination events. Here, we identify a large number of young de novo genes with discernible recent ancestral non-coding sequences and evidence of translation. Using pipelines examining the synteny relationship between genomes and reciprocal-best whole-genome alignments, we detected at least 175 de novo open reading frames in the focal species O. sativa subspecies japonica, which were all detected in RNA sequencing-based transcriptomes. Mass spectrometry-based targeted proteomics and ribosomal profiling show translational evidence for 57% of the de novo genes. In recent divergence of Oryza, an average of 51.5 de novo genes per million years were generated and retained. We observed evolutionary patterns in which excess indels and early transcription were favoured in origination with a stepwise formation of gene structure. These data reveal that de novo genes contribute to the rapid evolution of protein diversity under positive selection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chen, L., DeVries, A. L. & Cheng, C. H. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc. Natl Acad. Sci. USA 94, 3811–3816 (1997).
Levine, M. T., Jones, C. D., Kern, A. D., Lindfors, H. A. & Begun, D. J. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc. Natl Acad. Sci. USA 103, 9935–9939 (2006).
Ohno, S. Evolution by Gene Duplication (Springer, 1970).
Jacob, F. Evolution and tinkering. Science 196, 1161–1166 (1977).
Gilbert, W. Why genes in pieces? Nature 271, 501 (1978).
Mayr, E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance (Belknap Press, 1982).
Patthy, L. in Protein Evolution 2nd edn 108–109 (Blackwell Publishing, 2008).
Klasberg, S., Bitard-Feildel, T., Callebaut, I. & Bornberg-Bauer, E. Origins and structural properties of novel and de novo protein domains during insect evolution. FEBS J. 285, 2605–2625 (2018).
Bitard-Feildel, T., Heberlein, M., Bornberg-Bauer, E. & Callebaut, I. Detection of orphan domains in Drosophila using “hydrophobic cluster analysis”. Biochimie 119, 244–253 (2015).
Cai, J., Zhao, R., Jiang, H. & Wang, W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179, 487–496 (2008).
Carvunis, A. R. et al. Proto-genes and de novo gene birth. Nature 487, 370–374 (2012).
Xiao, W. et al. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 4, e4603 (2009).
Wu, D. D. et al. “Out of pollen” hypothesis for origin of new genes in flowering plants: study from Arabidopsis thaliana. Genome Biol. Evol. 6, 2822–2829 (2014).
Cui, X. et al. Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol. Plant 8, 935–945 (2015).
Donoghue, M. T., Keshavaiah, C., Swamidatta, S. H. & Spillane, C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 11, 47 (2011).
Begun, D. J., Lindfors, H. A., Kern, A. D. & Jones, C. D. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176, 1131–1137 (2007).
Chen, S. T., Cheng, H. C., Barbash, D. A. & Yang, H. P. Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster. PLoS Genet. 3, e107 (2007).
Chen, S., Zhang, Y. E. & Long, M. New genes in Drosophila quickly become essential. Science 330, 1682–1685 (2010).
Reinhardt, J. A. et al. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 9, e1003860 (2013).
Zhou, Q. et al. On the origin of new genes in Drosophila. Genome Res. 18, 1446–1455 (2008).
Zhao, L., Saelao, P., Jones, C. D. & Begun, D. J. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343, 769–772 (2014).
Toll-Riera, M. et al. Origin of primate orphan genes: a comparative genomics approach. Mol. Biol. Evol. 26, 603–612 (2009).
Li, C. Y. et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput. Biol. 6, e1000734 (2010).
Wu, D. D., Irwin, D. M. & Zhang, Y. P. De novo origin of human protein-coding genes. PLoS Genet. 7, e1002379 (2011).
Zhang, Y. E., Vibranovski, M. D., Landback, P., Marais, G. A. & Long, M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 8, e1000494 (2010).
Knowles, D. G. & McLysaght, A. Recent de novo origin of human protein-coding genes. Genome Res. 19, 1752–1759 (2009).
Murphy, D. N. & McLysaght, A. De novo origin of protein-coding genes in murine rodents. PLoS ONE 7, e48650 (2012).
Xie, C. et al. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 8, e1002942 (2012).
Ruiz-Orera, J., Verdaguer-Grau, P., Villanueva-Canas, J. L., Messeguer, X. & Alba, M. M. Translation of neutrally evolving peptides provides a basis for de novo gene evolution. Nat. Ecol. Evol. 2, 890–896 (2018).
Tautz, D. & Domazet-Lošo, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12, 692–702 (2011).
Schlötterer, C. Genes from scratch—the evolutionary fate of de novo genes. Trends Genet. 31, 215–219 (2015).
Moyers, B. A. & Zhang, J. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol. Biol. Evol. 33, 1245–1256 (2018).
Zhao, Y. et al. Identification and analysis of unitary loss of long-established protein-coding genes in Poaceae shows evidences for biased gene loss and putatively functional transcription of relics. BMC Evol. Biol. 15, 66 (2015).
Cheng, C. H. & Chen, L. Evolution of an antifreeze glycoprotein. Nature 401, 443–444 (1999).
Husnik, F. & McCutcheon, J. P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 16, 67–79 (2018).
Dujon, B. The yeast genome project: what did we learn? Trends Genet. 12, 263–270 (1996).
Gubala, A. M. et al. The goddard and saturn genes are essential for Drosophila male fertility and may have arisen de novo. Mol. Biol. Evol. 34, 1066–1082 (2017).
Stein, J. C. et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296 (2018).
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Sakai, H. et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6 (2013).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Long, M. Y., VanKuren, N. W., Chen, S. D. & Vibranovski, M. D. New gene evolution: little did we know. Annu. Rev. Genet. 47, 307–333 (2013).
Zhang, C. J. et al. High occurrence of functional new chimeric genes in survey of rice chromosome 3 short arm genome sequences. Genome Biol. Evol. 5, 1038–1048 (2013).
Zhang, Y. E., Landback, P., Vibranovski, M. & Long, M. New genes expressed in human brains: implications for annotating evolving genomes. BioEssays 34, 982–991 (2012).
Mills, R. E. et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16, 1182–1190 (2006).
Wang, W. et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49 (2018).
Xu, X. et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111 (2012).
Watterson, G. A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975).
McDonald, J. H. & Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991).
Wang, M. et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–988 (2014).
Hartl, D. L. & Clark, A. G. Principles of Population Genetics 4th edn 172–175; 351–354 (Sinauer Associates, Sunderland, 2007).
Berretta, J. & Morillon, A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10, 973–982 (2009).
Bornberg-Bauer, E. & Alba, M. M. Dynamics and adaptive benefits of modular protein evolution. Curr. Opin. Struct. Biol. 23, 459–466 (2013).
Neme, R., Amador, C., Yildirim, B., McConnell, E. & Tautz, D. Random sequences are an abundant source of bioactive RNAs or peptides. Nat. Ecol. Evol. 1, 0217 (2017).
Heinen, T. J., Staubach, F., Häming, D. & Tautz, D. Emergence of a new gene from an intergenic region. Curr. Biol. 19, 1527–1531 (2009).
Yanai, I. et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005).
Long, M., Rosenberg, C. & Gilbert, W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl Acad. Sci. USA 92, 12495–12499 (1995).
Sharp, P. A. Speculations on RNA splicing. Cell 23, 643–646 (1981).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Lange, V., Picotti, P., Domon, B. & Aebersold, R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222 (2008).
Ebhardt, H. A., Root, A., Sander, C. & Aebersold, R. Applications of targeted proteomics in systems biology and translational medicine. Proteomics 15, 3193–3208 (2015).
Pecorelli, I., Bibi, R., Fioroni, L. & Galarini, R. Validation of a confirmatory method for the determination of sulphonamides in muscle according to the European Union regulation 2002/657/EC. J. Chromatogr. A 1032, 23–29 (2004).
Wen, B. et al. IPeak: an open source tool to combine results from multiple MS/MS search engines. Proteomics 15, 2916–2920 (2015).
Zhao, D. et al. Analysis of ribosome-associated mRNAs in rice reveals the importance of transcript size and GC content in translation. G3 (Bethesda) 7, 203–219 (2017).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Sabi, R., Volvovitch Daniel, R. & Tuller, T. stAIcalc: tRNA adaptation index calculator based on species-specific weights. Bioinformatics 33, 589–591 (2017).
Lees, J. G., Dawson, N. L., Sillitoe, I. & Orengo, C. A. Functional innovation from changes in protein domains and their combinations. Curr. Opin. Struct. Biol. 38, 44–52 (2016).
Davidson, A. R. & Sauer, R. T. Folded proteins occur frequently in libraries of random amino acid sequences. Proc. Natl Acad. Sci. USA 91, 2146–2150 (1994).
Keefe, A. D. & Szostak, J. W. Functional proteins from a random-sequence library. Nature 410, 715–718 (2001).
Vaughan, D. A., Morishima, H. & Kadowaki, K. Diversity in the Oryza genus. Curr. Opin. Plant Biol. 6, 139–146 (2003).
Murat, F., Van de Peer, Y. & Salse, J. Decoding plant and animal genome plasticity from differential paleo-evolutionary patterns and processes. Genome Biol. Evol. 4, 917–928 (2012).
Huey, R. B. et al. Plants versus animals: do they deal with stress in different ways? Integr. Comp. Biol. 42, 415–423 (2002).
Wilson, B. A., Foy, S. G., Neme, R. & Masel, J. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat. Ecol. Evol. 1, 0146 (2017).
McLysaght, A. & Hurst, L. D. Open questions in the study of de novo genes: what, how and why. Nat. Rev. Genet. 17, 567–578 (2016).
Zhang, Y. E., Vibranovski, M. D., Krinsky, B. H. & Long, M. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20, 1526–1533 (2010).
Zhang, Y. E., Landback, P., Vibranovski, M. D. & Long, M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 9, e1001179 (2011).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Ranwez, V., Harispe, S., Delsuc, F. & Douzery, E. J. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS ONE 6, e22594 (2011).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, ii215–ii225 (2003).
Dos Reis, M. et al. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 32, 5036–5044 (2004).
Chan, P. P. & Lowe, T. M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009).
Aebersold, R., Burlingame, A. L. & Bradshaw, R. A. Western blots versus selected reaction monitoring assays: time to turn the tables? Mol. Cell. Proteomics 12, 2381–2382 (2013).
Sjostrom, M. et al. A combined shotgun and targeted mass spectrometry strategy for breast cancer biomarker discovery. J. Proteome Res. 14, 2807–2818 (2015).
Guo, J. et al. A comprehensive investigation toward the indicative proteins of bladder cancer in urine: from surveying cell secretomes to verifying urine proteins. J. Proteome Res. 15, 2164–2177 (2016).
Xie, Y. et al. The levels of serine proteases in colon tissue interstitial fluid and serum serve as an indicator of colorectal cancer progression. Oncotarget 7, 32592–32606 (2016).
Zhang, S. et al. Quantitative analysis of the human AKR family members in cancer cell lines using the mTRAQ/MRM approach. J. Proteome Res. 12, 2022–2033 (2013).
Hou, G. et al. Biomarker discovery and verification of esophageal squamous cell carcinoma using integration of SWATH/MRM. J. Proteome Res. 14, 3793–3803 (2015).
Hou, G., Wang, Y., Lou, X. & Liu, S. Combination strategy of quantitative proteomics uncovers the related proteins of colorectal cancer in the interstitial fluid of colonic tissue from the AOM-DSS mouse model. Methods Mol. Biol. 1788, 185–192 (2017).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 (2014).
Uhlen, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Lindskog, C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev. Proteomics 12, 213–215 (2015).
Uhlen, M. et al. Transcriptomics resources of human tissues and organs. Mol. Syst. Biol. 12, 862 (2016).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Picotti, P. & Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 (2012).
Reiter, L. et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 (2011).
Bruderer, R., Bernhardt, O. M., Gandhi, T. & Reiter, L. High-precision iRT prediction in the targeted analysis of data-independent acquisition and its impact on identification and quantitation. Proteomics 16, 2246–2256 (2016).
Navarro, P. et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol. 34, 1130–1136 (2016).
Jordan, G. & Goldman, N. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol. Biol. Evol. 29, 1125–1139 (2012).
Löytynoja, A. Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170 (2014).
Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573 (1998).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Acknowledgements
We appreciate valuable discussions with N. Jiang at MSU, the group of M. L. at Chicago, Y. Liao and M. Chen at the Institute of Genetics and Development in Beijing, and J. P. Staley at Chicago. We are thankful for the editing done by E. Mortola. This work was supported by the USA National Science Foundation (NSF) under Plant Genome Research Program numbers 0321678, 0638541 and 0822284, the Bud Antle Endowed Chair of Excellence in Agriculture and Life Sciences, and the AXA Chair for Evolutionary Genomics and Genome Biology (to R.A.W.), NSF MCB number 1026200 (to M.L. and R.A.W.), NSF MCB 1051826 and NIH R01 GM 100768 (to M.L.), the National Key R&D Program of China 2017YFC0908400 (to S.L.) and the National Program for Support of Top-notch Young Professionals of China (to Y.O.).
Author information
Authors and Affiliations
Contributions
L.Z., R.A.W., S.L. and M.L. conceived and designed the project. L.Z., Y.R., R.A.W., S.L. and M.L. wrote the manuscript, with significant contributions from C.Z., A.R.G., J.C. and Y.Z. L.Z. conducted the computational genomic analysis, with significant contributions from A.R.G., K.C., J.Z. and Y.Z. C.Z., Y.Y., J.Z., K.C., M.W., D.C. and R.A.W. generated and annotated the genome sequences. Y.R., G.H., J.Z., L.Z. and S.L. designed and conducted the proteomics experiments to detect proteins translated from de novo genes. R.Z., B.W., L.Z. and Z.P. conducted the analysis of public proteomics databases. Y.R., L.Z., J.C., M.L. and S.L. performed further evolutionary and proteomics analyses. T.Y., G.L. and Y.O. grew rice strains in Sanya (China) and dissected rice tissues. J.C., L.Z., C.Z. and M.L. conducted the evolutionary substitution analyses of de novo genes.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Figures
Supplementary Figures 1–6
Supplementary File 1
Sequence alignments of 929 orphan genes exported from the MASCE program. Alignments were manually annotated at a later stage and can be found online
Supplementary File 2
Ribosome profiling evidence for candidate de novo genes
Supplementary Table 1
ORF status of de novo gene candidates in each species
Supplementary Table 2
Transcription status of de novo gene candidates in each species
Supplementary Table 3
Candidate de novo genes with matches in the Genbank’s nr database
Supplementary Table 4
Statistics of mutations that are crucial for the transformation of noncoding to coding sequences
Supplementary Table 5
Population genomics of indels and SNP in O. sativa japonica and O. barthii.
Supplementary Table 6
Expression level and tissue specificity of candidate de novo genes and old singleton 76 genes derived from OGE datasets including leaf, root, and panicle.
Supplementary Table 7
Gene structures with relevant statistics
Supplementary Table 8
Intron phase distributions for different gene categories
Supplementary Table 9
Candidate de novo genes with signals of natural selection resulting from the branch model analyses in PAML
Supplementary Table 10
. Candidate de novo genes that have been identified with peptide supports by the MRM method.
Supplementary Table 11
The eight datasets used for proteomics analysis of candidate de novo genes
Supplementary Table 12
Candidate de novo genes that have been identified with peptide supports.
Supplementary Table 13
Candidate de novo genes with ribosomal profiling evidence supports.
Supplementary Table 14
tRNA adaptive indexes (tAIs) in 175 de novo genes (plus 7 isoforms) and 4,965 single-copy genes (plus 2,079 isoforms).
Rights and permissions
About this article
Cite this article
Zhang, L., Ren, Y., Yang, T. et al. Rapid evolution of protein diversity by de novo origination in Oryza. Nat Ecol Evol 3, 679–690 (2019). https://doi.org/10.1038/s41559-019-0822-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0822-5
This article is cited by
-
The origin and structural evolution of de novo genes in Drosophila
Nature Communications (2024)
-
Four classic “de novo” genes all have plausible homologs and likely evolved from retro-duplicated or pseudogenic sequences
Molecular Genetics and Genomics (2024)
-
Clustering pattern and evolution characteristic of microRNAs in grass carp (Ctenopharyngodon idella)
BMC Genomics (2023)
-
Uncovering gene-family founder events during major evolutionary transitions in animals, plants and fungi using GenEra
Genome Biology (2023)
-
Selection of a de novo gene that can promote survival of Escherichia coli by modulating protein homeostasis pathways
Nature Ecology & Evolution (2023)