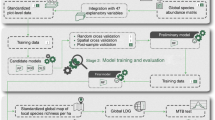
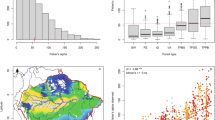
Abstract
Controversy remains over what drives patterns in the variation of biodiversity across the planet. The resolution is obscured by lack of data and mismatches in their spatial grain (scale), and by grain-dependent effects of the drivers. Here we introduce cross-scale models integrating global data on tree-species richness from 1,336 local forest surveys and 282 regional checklists, enabling the estimation of drivers and patterns of biodiversity across spatial grains. We uncover grain-dependent effects of both environment and biogeographic regions on species richness, with a striking positive effect of Southeast Asia at coarse grain that disappears at fine grains. We show that, globally, biodiversity cannot be attributed purely to environmental or regional drivers, as the regions are environmentally distinct even within a single latitudinal band. Finally, we predict global maps of biodiversity at local (plot-based) and regional grains, identifying areas of exceptional beta-diversity in China, East Africa and North America. By allowing the importance of drivers of diversity to vary with grain in a single model, our approach unifies disparate results from previous studies regarding environmental versus biogeographic predictors of biodiversity, and enables efficient integration of heterogeneous data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data and R codes used for the analyses are available under CC-BY 4.0 license in a GitHub repository at https://github.com/petrkeil/global_tree_S, which is also mirrored at figshare at https://figshare.com/articles/global_tree_S/7461509. Please note that if the data on species richness are reused, the original data sources should be credited.
References
Fine, P. V. A. & Ree, R. H. Evidence for a time-integrated species-area effect on the latitudinal gradient in tree diversity. Am. Nat. 168, 796–804 (2006).
Frodin, D. G. Guide to Standard Floras of the World (Cambridge Univ. Press, Cambridge, 2001).
Losos, E. & Leigh, E. G. Tropical Forest Diversity and Dynamism (Univ. of Chicago Press, Chicago, 2004).
Hawkins, B. A. Ecology’s oldest pattern? Trends Ecol. Evol. 16, 470 (2001).
Storch, D., Bohdalková, E. & Okie, J. The more-individuals hypothesis revisited: the role of community abundance in species richness regulation and the productivity-diversity relationship. Ecol. Lett. 21, 920–937 (2018).
Currie, D. J. et al. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol. Lett. 7, 1121–1134 (2004).
Ricklefs, R. E. & He, F. Region effects influence local tree species diversity. Proc. Natl Acad. Sci. USA 113, 674–679 (2016).
Wiens, J. J. et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324 (2010).
Rabosky, D. L. & Hurlbert, A. H. Species richness at continental scales is dominated by ecological limits. Am. Nat. 185, 572–583 (2015).
Meyer, C., Kreft, H., Guralnick, R. & Jetz, W. Global priorities for an effective information basis of biodiversity distributions. Nat. Commun. 6, 8221 (2015).
Jetz, W., McPherson, J. M. & Guralnick, R. P. Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159 (2012).
Phillips, O. L. & Miller, J. S. Global Patterns of Plant Piversity: Alwyn H. Gentry’s Forest Transect Data Set (Missouri Botanical Garden Press, St. Louis, 2002).
Sullivan, M. et al. Diversity and carbon storage across the tropical forest biome. Sci. Rep. 7, 39102 (2017).
GlobalTreeSearch Online Database (BCGI, 2017); https://www.bgci.org/global_tree_search.php
Enquist, B. J., Condit, R., Peet, R. K., Schildhauer, M. & Thiers, B. M. Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Preprints 4, e2615v2 (2016).
Levin, S. A. Multiple scales and the maintenance of biodiversity. Ecosystems 3, 498–506 (2000).
Chave, J. The problem of pattern and scale in ecology: what have we learned in 20 years? Ecol. Lett. 16, 4–16 (2013).
Chase, J. M. Spatial scale resolves the niche versus neutral theory debate. J. Veg. Sci. 25, 319–322 (2014).
Leibold, M. A. & Chase, J. M. Metacommunity Ecology (Princeton Univ. Press, Princeton, 2017).
Storch, D. The theory of the nested species–area relationship: geometric foundations of biodiversity scaling. J. Veg. Sci. 27, 880–891 (2016).
Rahbek, C. The role of spatial scale and the perception of large-scale species-richness patterns. Ecol. Lett. 8, 224 (2005).
Rahbek, C. & Graves, G. R. Detection of macro-ecological patterns in South American hummingbirds is affected by spatial scale. Proc. R. Soc. B 267, 2259–2265 (2000).
Chase, J. M. & Knight, T. M. Scale-dependent effect sizes of ecological drivers on biodiversity: why standardised sampling is not enough. Ecol. Lett. 16, 17–26 (2013).
Blowes, S. A., Belmaker, J. & Chase, J. M. Global reef fish richness gradients emerge from divergent and scale-dependent component changes. Proc. R. Soc. B 284, 20170947 (2017).
Kraft, N. J. B. et al. Disentangling the drivers of β diversity along latitudinal and elevational gradients. Science 333, 1755–1758 (2011).
Buckley, L. B. & Jetz, W. Linking global turnover of species and environments. Proc. Natl Acad. Sci. USA 105, 17836–17841 (2008).
Shmida, A. & Wilson, M. V. Biological determinants of species diversity. J. Biogeogr. 12, 1–20 (1985).
Böhning-Gaese, K. Determinants of avian species richness at different spatial scales. J. Biogeogr. 24, 49–60 (1997).
Belmaker, J. & Jetz, W. Cross-scale variation in species richness–environment associations. Glob. Ecol. Biogeogr. 20, 464–474 (2011).
Rahbek, C. & Graves, G. R. Multiscale assessment of patterns of avian species richness. Proc. Natl Acad. Sci. USA 98, 4534–4539 (2001).
Wang, Z., Brown, J. H., Tang, Z. & Fang, J. Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl Acad. Sci. USA 106, 13388–13392 (2009).
Whittaker, R. J., Willis, K. J. & Field, R. Scale and species richness: towards a general, hierarchical theory of species diversity. J. Biogeogr. 28, 453–470 (2001).
Ricklefs, R. E. Intrinsic dynamics of the regional community. Ecol. Lett. 18, 497–503 (2015).
Vázquez-Rivera, H. & Currie, D. J. Contemporaneous climate directly controls broad-scale patterns of woody plant diversity: a test by a natural experiment over 14,000 years. Glob. Ecol. Biogeogr. 24, 97–106 (2015).
Fine, P. V. A. Ecological and evolutionary drivers of geographic variation in species diversity. Annu. Rev. Ecol. Evol. Syst. 46, 369–392 (2015).
Harmon, L. J. & Harrison, S. Species diversity is dynamic and unbounded at local and continental scales. Am. Nat. 185, 584–593 (2015).
Wiens, J. J., Pyron, R. A. & Moen, D. S. Phylogenetic origins of local-scale diversity patterns and the causes of Amazonian megadiversity. Ecol. Lett. 14, 643–652 (2011).
Hawkins, B. A., Porter, E. E. & Diniz-Filho, J. A. F. Productivity and history as predictors of the latitudinal diversity gradient of terrestrial birds. Ecology 84, 1608–1623 (2003).
Algar, A. C., Kerr, J. T. & Currie, D. J. Evolutionary constraints on regional faunas: whom, but not how many. Ecol. Lett. 12, 57–65 (2009).
Dunn, R. R. et al. Climatic drivers of hemispheric asymmetry in global patterns of ant species richness. Ecol. Lett. 12, 324–333 (2009).
Araújo, M. B. et al. Quaternary climate changes explain diversity among reptiles and amphibians. Ecography 31, 8–15 (2008).
Belmaker, J. & Jetz, W. Relative roles of ecological and energetic constraints, diversification rates and region history on global species richness gradients. Ecol. Lett. 18, 563–571 (2015).
Kreft, H. & Jetz, W. Global patterns and determinants of vascular plant diversity. Proc. Natl Acad. Sci. U.S.A. 104, 5925–5930 (2007).
Currie, D. J. & Paquin, V. Large-scale biogeographical patterns of species richness of trees. Nature 329, 326 (1987).
Francis, A. P. & Currie, D. J. Global patterns of tree species richness in moist forests: another look. Oikos 81, 598–602 (1998).
Šímová, I. et al. Global species–energy relationship in forest plots: role of abundance, temperature and species climatic tolerances. Glob. Ecol. Biogeogr. 20, 842–856 (2011).
Latham, R. & Ricklefs, R. E. Global patterns of tree species richness in moist forests: energy-diversity theory does not account for variation in species richness. Oikos 67, 325–333 (1993).
Ricklefs, R. E., Latham, R. E. & Qian, H. Global patterns of tree species richness in moist forests: distinguishing ecological influences and historical contingency. Oikos 86, 369–373 (1999).
Qian, H., Wiens, J. J., Zhang, J. & Zhang, Y. Evolutionary and ecological causes of species richness patterns in North American angiosperm trees. Ecography 38, 241–250 (2015).
Qian, H. & Ricklefs, R. E. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182 (2000).
Ricklefs, R. E., Qian, H. & White, P. S. The region effect on mesoscale plant species richness between eastern Asia and eastern North America. Ecography 27, 129–136 (2004).
Lyons, S. K. & Willig, M. R. A hemispheric assessment of scale dependence in latitudinal gradients of species richness. Ecology 80, 2483–2491 (1999).
O’Brien, E. M., Field, R. & Whittaker, R. J. Climatic gradients in woody plant (tree and shrub) diversity: water-energy dynamics, residual variation, and topography. Oikos 89, 588–600 (2000).
Field, R., O’Brien, E. M. & Whittaker, R. J. Global models for predicting woody plant richness from climate: development and evaluation. Ecology 86, 2263–2277 (2005).
Brown, J. H. Macroecology (Univ. of Chicago Press, Chicago, 1995).
Mutke, J. & Barthlott, W. Patterns of vascular plant diversity at continental to global scale. Biol. Skrift. 55, 521–538 (2005).
Lomolino, M. V., Riddle, B. R., Whittaker, R. J. & Brown, J. H. Biogeography (Sinauer Associates, Sunderland, 2010).
Qian, H. A comparison of the taxonomic richness of temperate plants in East Asia and North America. Am. J. Bot. 89, 1818–1825 (2002).
Crist, T. O. & Veech, J. A. Additive partitioning of rarefaction curves and species-area relationships: unifying alpha-, beta- and gamma-diversity with sample size and habitat area. Ecol. Lett. 9, 923–932 (2006).
Svenning, J.-C. & Skov, F. Limited filling of the potential range in European tree species: limited range filling in European trees. Ecol. Lett. 7, 565–573 (2004).
Jansson, R. & Davies, T. J. Global variation in diversification rates of flowering plants: energy vs. climate change. Ecol. Lett. 11, 173–183 (2007).
Legendre, P. Spatial autocorrelation: trouble or new paradigm? Ecology 74, 1659–1673 (1993).
Dormann, C. F. et al. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628 (2007).
Dormann, C. F. Effects of incorporating spatial autocorrelation into the analysis of species distribution data. Glob. Ecol. Biogeogr. 16, 129–138 (2007).
Quintero, I. & Jetz, W. Global elevational diversity and diversification of birds. Nature 555, 246–250 (2018).
Stein, A., Gerstner, K. & Kreft, H. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol. Lett. 17, 866–880 (2014).
MacArthur, R. H. & Wilson, E. O. The Theory of Island Biogeography (Princeton Univ. Press, Princeton, 1967).
Carrete, M. et al. Habitat, human pressure, and social behavior: Partialling out factors affecting large-scale territory extinction in an endangered vulture. Biol. Conserv. 136, 143–154 (2007).
Randin, C. F. et al. Climate change and plant distribution: local models predict high‐elevation persistence. Glob. Change Biol. 15, 1557–1569 (2009).
White, E. P. & Hurlbert, A. H. The combined influence of the local environment and regional enrichment on bird species richness. Am. Nat. 175, E35–E43 (2010).
Dormann, C. F. et al. co-linearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36, 27–46 (2013).
Rominger, A. J. et al. Community assembly on isolated islands: macroecology meets evolution. Glob. Ecol. Biogeogr. 25, 769–780 (2016).
Swenson, N. G. et al. Constancy in functional space across a species richness anomaly. Am. Nat. 187, E83–E92 (2016).
Šizling, A. L. et al. Can people change the ecological rules that appear general across space? Glob. Ecol. Biogeogr. 25, 1072–1084 (2016).
Quintero, I., Keil, P., Jetz, W. & Crawford, F. W. Historical biogeography using species geographical ranges. Syst. Biol. 64, 1059–1073 (2015).
Arias, J. S. An event model for phylogenetic biogeography using explicitly geographical ranges. J. Biogeogr. 44, 2225–2235 (2017).
Hawkins, B. A. & Porter, E. E. Relative influences of current and historical factors on mammal and bird diversity patterns in deglaciated North America: climate, ice and diversity. Glob. Ecol. Biogeogr. 12, 475–481 (2003).
Sandel, B. et al. The influence of late quaternary climate-change velocity on species endemism. Science 334, 660–664 (2011).
Jetz, W. & Fine, P. V. A. Global gradients in vertebrate diversity predicted by historical area-productivity dynamics and contemporary environment. PLoS Biol. 10, e1001292 (2012).
Cabral, J. S., Valente, L. & Hartig, F. Mechanistic simulation models in macroecology and biogeography: state-of-art and prospects. Ecography 40, 267–280 (2017).
Connolly, S. R., Keith, S. A., Colwell, R. K. & Rahbek, C. Process, mechanism, and modeling in macroecology. Trends Ecol. Evol. 32, 835–844 (2017).
McGill, B. Strong and weak tests of macroecological theory. Oikos 102, 679–685 (2003).
Coelho de Souza, F. et al. Evolutionary heritage influences Amazon tree ecology. Proc. R. Soc. B 283, 20161587 (2016).
Phillips, O. L. et al. Efficient plot-based floristic assessment of tropical forests. J. Trop. Ecol. 19, 629–645 (2003).
Ramesh, B. R. et al. Forest stand structure and composition in 96 sites along environmental gradients in the central Western Ghats of India. Ecology 91, 3118–3118 (2010).
Myers, J. A., Chase, J. M., Crandall, R. M. & Jiménez, I. Disturbance alters beta-diversity but not the relative importance of community assembly mechanisms. J. Ecol. 103, 1291–1299 (2015).
US Department of Agriculture. Forest Inventory and Analysis – Fiscal Year 2016 Business Report (US Department of Agriculture, Washington, D.C., 2016).
De Natale, F. et al. Inventario Nazionale delle Foreste e dei Serbatoi Forestali di Carbonio (Ispettorato Generale del Corpo Forestale dello Stato, CRA-ISAFA, Trento, 2005).
Institut national de l’information géographique et forestière. French National Forest Inventory (FNFI) (IGN, Saint-Mandé, 2017); http://inventaire-forestier.ign.fr/
Abbott, I. Comparisons of spatial pattern, structure, and tree composition between virgin and cut-over jarrah forest in Western Australia. For. Ecol. Manag. 9, 101–126 (1984).
Adam, J. H. Changes in forest community structures of tropical montane rain forest on the slope of Mt. Trus Madi in Sabah, Malaysia. J. Trop. For. Sci. 13, 76–92 (2001).
Addo-Fordjour, P., Obeng, S., Anning, A. & Addo, M. Floristic composition, structure and natural regeneration in a moist semi-deciduous forest following anthropogenic disturbances and plant invasion. Int. J. Biodiv. Conserv. 1, 21–37 (2009).
Adekunle, V. A. J. Conservation of tree species diversity in tropical rainforest ecosystem of South-West Nigeria. J. Trop. For. Sci. 18, 91–101 (2006).
Ansley, S. J.-A. & Battles, J. J. Forest composition, structure, and change in an old-growth mixed conifer forest in the northern Sierra Nevada. J. Torrey Bot. Soc. 125, 297–308 (1998).
Beals, E. W. The remnant cedar forests of Lebanon. J. Ecol. 53, 679–694 (1965).
Bonino, E. E. & Araujo, P. Structural differences between a primary and a secondary forest in the Argentine Dry Chaco and management implications. For. Ecol. Manag. 206, 407–412 (2005).
Cairns, M. A., Olmsted, I., Granados, J. & Argaez, J. Composition and aboveground tree biomass of a dry semi-evergreen forest on Mexico’s Yucatan Peninsula. For. Ecol. Manag. 186, 125–132 (2003).
Cao, M. & Zhang, J. Tree species diversity of tropical forest vegetation in Xishuangbanna, SW China. Biodivers. Conserv. 6, 995–1006 (1997).
Cheng-Yang, Z., Zeng-Li, L. I. U. & Jing-Yun, F. Tree species diversity along latitudinal gradient on southeastern and northwestern slopes of Mt. Huanggang, Wuyi Mountains, Fujian, China. Biodivers. Sci. 12, 63–74 (2004).
Davis, M. A., Curran, C., Tietmeyer, A. & Miller, A. Dynamic tree aggregation patterns in a species-poor temperate woodland disturbed by fire. J. Veg. Sci. 16, 167–174 (2005).
Do, T. V. et al. Effects of micro-topographies on stand structure and tree species diversity in an old-growth evergreen broad-leaved forest, southwestern Japan. Glob. Ecol. Conserv. 4, 185–196 (2015).
Eichhorn, M. Boreal forests of Kamchatka: structure and composition. Forests 1, 154–176 (2010).
Enoki, T. Microtopography and distribution of canopy trees in a subtropical evergreen broad-leaved forest in the northern part of Okinawa Island, Japan. Ecol. Res. 18, 103–113 (2003).
Eshete, A., Sterck, F. & Bongers, F. Diversity and production of Ethiopian dry woodlands explained by climate- and soil-stress gradients. For. Ecol. Manag. 261, 1499–1509 (2011).
Fashing, P. J., Forrestel, A., Scully, C. & Cords, M. Long-term tree population dynamics and their implications for the conservation of the Kakamega Forest, Kenya. Biodivers. Conserv. 13, 753–771 (2004).
Graham, A. W. The CSIRO Rainforest Permanent Plots of North Queensland - Site, Structural, Floristic and Edaphic Descriptions (CSIRO and the Cooperative Research Centre for Tropical Rainforest Ecology and Management, Rainforest CRC, Cairns, 2006).
Hirayama, K. & Sakimoto, M. Spatial distribution of canopy and subcanopy species along a sloping topography in a cool‐temperate conifer‐hardwood forest in the snowy region of Japan. Ecol. Res. 18, 443–454 (2003).
Jing-Yun, F., Yi-De, L. I., Biao, Z. H. U., Guo-Hua, L. I. U. & Guang-Yi, Z. Community structures and species richness in the montane rain forest of Jianfengling, Hainan Island, China. Biodivers. Sci. 12, 29–43 (2004).
Kohira, M., Ninomiya, I., Ibrahim, A. Z. & Latiff, A. Diversity, diameter structure and spatial pattern of trees in semi-evergreen rain forest of Langkawi island, Malaysia. J. Trop. For. Sci. 13, 460–476 (2001).
Kohyama, T. Tree size structure of stands and each species in primary warm-temperate rain forests of Southern Japan. Bot. Mag. Tokyo 99, 267–279 (1986).
Krishnamurthy, Y. L. et al. Vegetation structure and floristic composition of a tropical dry deciduous forest in Bhadra Wildlife Sanctuary, Karnataka, India. Trop. Ecol. 51, 235–246 (2010).
Lalfakawma, Sahoo, U., Roy, S., Vanlalhriatpuia, K. & Vanalalhluna, P. C. Community composition and tree population structure in undisturbed and disturbed tropical semi-evergreen forest stands of North-East India. Appl. Ecol. Env. Res. 7, 303–318 (2010).
Linder, P., Elfving, B. & Zackrisson, O. Stand structure and successional trends in virgin boreal forest reserves in Sweden. For. Ecol. Manag. 98, 17–33 (1997).
Lopes, C. G. R., Ferraz, E. M. N. & Araújo, EdeL. Physiognomic-structural characterization of dry- and humid-forest fragments (Atlantic Coastal Forest) in Pernambuco State, NE Brazil. Plant Ecol. 198, 1–18 (2008).
Lü, X.-T., Yin, J. & Tang, J.-W. Structure, tree species diversity and composition of tropical seasonal rainforests in Xishuangbanna, South-West China. J. Trop. For. Sci. 22, 260–270 (2010).
Malizia, A. & Grau, R. Liana–host tree associations in a subtropical montane forest of north-western Argentina. J. Trop. Ecol. 22, 331–339 (2006).
Maycock, F. P., Guzik, J., Jankovic, J., Shevera, M. & Carleton, J. T. Composition, structure and ecological aspects of mesic old growth Carpathian deciduous forests of Slovakia, southern Poland and the western Ukraine. Fragm. Flor. Geobot. 45, 281–321 (2000).
Nagel, A. T., Svoboda, M., Rugani, T. & Diaci, J. Gap regeneration and replacement patterns in an old-growth Fagus–Abies forest of Bosnia–Herzegovina. Plant. Ecol. 208, 307–318 (2010).
Namikawa, K., Matsui, T., Kobayashi, M., Goto, R. & Kuramoto, S. Initial establishment and regeneration processes of an outlying isolated Fagus crenata Blume forest stand in the northernmost boundary of its range in Hokkaido, northern Japan. Plant. Ecol. 207, 161–174 (2010).
Narayanan, A. & Parthasarathy, N. Biodiversity inventory of trees in a large scale permanent plot of tropical evergreen forest at Varagaliar. Anamalais, Western Ghats, India. Biodivers. Conserv. 8, 1533–1554 (1999).
Popradit, A. et al. Anthropogenic effects on a tropical forest according to the distance from human settlements. Sci. Rep. 5, 14689 (2015).
Round, P., Pierce, A., Sankamethawee, W. & Gale, G. The Mo Singto forest dynamics plot, Khao Yai National Park, Thailand. Nat. Hist. Bull. Siam Soc. 57, 57–80 (2011).
Sanchez, M., Pedroni, F., Eisenlohr, P. V. & Oliveira-Filho, A. T. Changes in tree community composition and structure of Atlantic rain forest on a slope of the Serra do Mar range, southeastern Brazil, from near sea level to 1000m of altitude. Flora 208, 184–196 (2013).
Sawada, H., Ohkubo, T., Kaji, M. & Oomura, K. Spatial distribution and topographic dependence of vegetation types and tree populations of natural forests in the Chichibu Mountains, central Japan. J. Japan. Forest Soc. 87, 293–303 (2005).
Sheil, D. & Salim, A. Forest tree persistence, elephants, and stem scars. Biotropica 36, 505–521 (2004).
Shu-Qing, Z. et al. Structure and species diversity of boreal forests in Mt. baikalu, huzhong area, daxing’an mountains, northeast china. Biodivers. Sci. 12, 182–189 (2004).
Splechtna, B. E., Gratzer, G. & Black, B. A. Disturbance history of a European old-growth mixed-species forest—A spatial dendro-ecological analysis. J. Veg. Sci. 16, 511–522 (2005).
Szwagrzyk, J. & Gazda, A. Above-ground standing biomass and tree species diversity in natural stands of Central Europe. J. Veg. Sci. 18, 555–562 (2007).
Wu, X.-P., Zhu, B. & Zhao, S.-Q. Comparison of community structure and species diversity of mixed forests of deciduous broad-leaved tree and Korean pine in Northeast China. Biodivers. Sci. 12, 174–181 (2004).
Wusheng, X., Tao, D., Shihong, L. & Li, X. A comparison of tree species diversity in two subtropical forests, Guangxi, Southwest China. J. Res. Ecol. 6, 208–216 (2015).
Yamada, I. Forest ecological studies of the montane forest of Mt. Pangrango, West Java: II. Stratification and floristic composition of the forest vegetation of the higher part of Mt. Pangrango. South East Asian Studies 13, 513–534 (1976).
Yasuoka, H. The variety of forest vegetations in south-eastern Cameroon, with special reference to the availability of wild yams for the forest hunter-gatherers. Afr. Study Monogr. 30, 89–119 (2008).
Kartesz, J. T. The Biota of North America Program (BONAP) (North American Plant Atlas, Chapel Hill, 2015).
Qian, H. Environmental determinants of woody plant diversity at a regional scale in China. PLoS. ONE 8, e75832 (2013).
Forzza, R. C. et al. Flora do Brazil 2020 (Jardim Botânico do Rio de Janeiro, Rio de Janeiro, 2017); http://floradobrasil.jbrj.gov.br/
Liang, J. et al. Positive biodiversity-productivity relationship predominant in global forests. Science 354, aaf8957 (2016).
Wood, S. N. Generalized Additive Models: an Introduction with R (CRC Press/Taylor & Francis Group, 2017).
Carpenter, B. et al. Stan: a probabilistic programming language. J. Stat. Softw. 76, https://doi.org/10.18637/jss.v076.i01 (2017).
Bürkner, P.-C. brms: an R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, https://doi.org/10.18637/jss.v080.i01 (2017).
Dauby, G. et al. RAINBIO: a mega-database of tropical African vascular plants distributions. PhytoKeys 74, 1–18 (2016).
Anderson-Teixeira, K. J. et al. CTFS-ForestGEO: a worldwide network monitoring forests in an era of global change. Glob. Change Biol. 21, 528–549 (2015).
DeWalt, S. J., Bourdy, G., Chavez de Michel, L. R. & Quenevo, C. Ethnobotany of the Tacana: Quantitative inventories of two permanent plots of Northwestern Bolivia. Econ. Bot. 53, 237–260 (1999).
Enquist, B. & Boyle, B. SALVIAS—the SALVIAS vegetation inventory database. Biodivers. Ecol. 4, 288–288 (2012).
Fegraus, E. Tropical ecology assessment and monitoring network (TEAMNetwork). Biodivers. Ecol. 4, 287–287 (2012).
Oliveira-filho, A. T. NeoTropTree, Flora Arbórea da Regiāo Neotropical: um Banco de Dados Envolvendo Biogeografia, Diversidade e Conservaçāo (Universidade Federal de Minas Gerais, Belo Horizonte, 2017).
Peet, R. K. et al. Vegetation-plot database of the Carolina Vegetation Survey. Biodivers. Ecol. 4, 243–253 (2012).
Peet, R. K., Lee, M. T., Jennings, M. D. & Faber-Langendoen, D. VegBank: a permanent, open-access archive for vegetation plot data. Biodivers. Ecol. 4, 233–241 (2012).
Sosef, M. S. M. et al. Exploring the floristic diversity of tropical Africa. BMC Biol. 15, 15 (2017).
Canhos, V. P. et al. Rede speciesLink: avaliação 2006 (Fapesp, São Paulo, 2006); http://splink.cria.org.br
Maitner, B. S. et al. The BIEN R package: A tool to access the Botanical Information and Ecology Network (BIEN)database. Meth. Eco. Evo. 9, 373–379 (2018).
Mauri, A., Strona, G. & San-Miguel-Ayanz, J. EU-Forest, a high-resolution tree occurrence dataset for Europe. Sci. Data 4, 160123 (2017).
Borcard, D., Legendre, P. & Drapeau, P. Partialling out the spatial component of ecological variation. Ecology 73, 1045–1055 (1992).
Acknowledgements
We thank D. Craven and I. Šímová for valuable advice, and H. Kreft, J. Coyle, R. Ricklefs, and S. Blowes for critical comments that greatly improved the manuscript. We acknowledge the support of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig funded by the German Research Foundation (FZT 118).
Author information
Authors and Affiliations
Contributions
P.K. formalized the ideas, collated the data, performed the analyses, and led the writing. J.M.C. proposed the initial idea, contributed to its development, discussed the results, and contributed to the writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–13, Supplementary Table 1, Supplementary Methods, Supplementary Discussion and Supplementary References
Rights and permissions
About this article
Cite this article
Keil, P., Chase, J.M. Global patterns and drivers of tree diversity integrated across a continuum of spatial grains. Nat Ecol Evol 3, 390–399 (2019). https://doi.org/10.1038/s41559-019-0799-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0799-0
This article is cited by
-
Latitudinal patterns in stabilizing density dependence of forest communities
Nature (2024)
-
Climate change and land use threaten global hotspots of phylogenetic endemism for trees
Nature Communications (2023)
-
Integrating remote sensing with ecology and evolution to advance biodiversity conservation
Nature Ecology & Evolution (2022)
-
Global patterns of vascular plant alpha diversity
Nature Communications (2022)
-
In memoriam Ching-I Peng (1950–2018)—an outstanding scientist and mentor with a remarkable legacy
Botanical Studies (2020)