Abstract
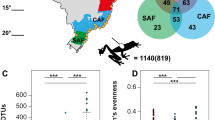
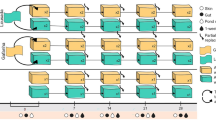
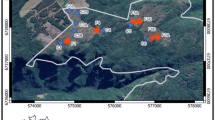
Animal-associated microbiomes are integral to host health, yet key biotic and abiotic factors that shape host-associated microbial communities at the global scale remain poorly understood. We investigated global patterns in amphibian skin bacterial communities, incorporating samples from 2,349 individuals representing 205 amphibian species across a broad biogeographic range. We analysed how biotic and abiotic factors correlate with skin microbial communities using multiple statistical approaches. Global amphibian skin bacterial richness was consistently correlated with temperature-associated factors. We found more diverse skin microbiomes in environments with colder winters and less stable thermal conditions compared with environments with warm winters and less annual temperature variation. We used bioinformatically predicted bacterial growth rates, dormancy genes and antibiotic synthesis genes, as well as inferred bacterial thermal growth optima to propose mechanistic hypotheses that may explain the observed patterns. We conclude that temporal and spatial characteristics of the host’s macro-environment mediate microbial diversity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
A full description of data analyses is provided in the Supplementary Information. Data for all newly sequenced samples is available on the Short Read Archive (Bioproject PRJNA474496). All figures include associated raw data and there are no restrictions on data availability. Correspondence and requests for materials should be addressed to M.V. or D.C.W.
References
Thompson, L. et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Hird, S. M. Evolutionary biology needs wild microbiomes. Front. Microbiol. 8, 1–10 (2017).
Jiménez, R. R. & Sommer, S. The amphibian microbiome: natural range of variation, pathogenic dysbiosis, and role in conservation. Biodivers. Conserv. 26, 763–786 (2016).
Lear, G. et al. Following Rapoport’s Rule: the geographic range and genome size of bacterial taxa decline at warmer latitudes. Environ. Microbiol. 8, 3152–3162 (2017).
Amend, A. S. et al. Macroecological patterns of marine bacteria on a global scale. J. Biogeogr. 40, 800–811 (2013).
Baldwin, A. J. et al. Microbial diversity in a pacific ocean transect from the arctic to antarctic circles. Aquat. Microb. Ecol. 41, 91–102 (2005).
Fuhrman, J. A. et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl Acad. Sci. USA 105, 7774–7778 (2008).
Tedersoo, L. & Nara, K. General latitudinal gradient of biodiversity is reversed in ectomycorrhizal fungi. New Phytol. 185, 351–354 (2010).
Ladau, J. et al. Global marine bacterial diversity peaks at high latitudes in winter. ISME. J. 7, 1669–1677 (2013).
Milici, M. et al. Low diversity of planktonic bacteria in the tropical ocean. Sci. Rep. 6, 19054 (2016).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl Acad. Sci. USA 103, 626–631 (2006).
Lozupone, C. A. & Knight, R. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA 104, 11436–11440 (2007).
Jones, S. E. & Lennon, J. T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl Acad. Sci. USA 107, 5881–5886 (2010).
Crevecoeur, S., Vincent, W. F., Comte, J. & Lovejoy, C. Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front. Microbiol. 6, 1–15 (2015).
Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2012).
Bletz, M. C. et al. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820 (2013).
Walke, J. B. & Belden, L. K. Harnessing the microbiome to prevent fungal infections: lessons from amphibians. PLoS Pathog. 12, e1005796 (2016).
Fisher, M. C., Garner, T. W. & Walker, S. F. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu. Rev. Microbiol. 63, 291–310 (2009).
Lips, K. R. et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc. Natl Acad. Sci. USA 103, 3165–3170 (2006).
Martel, A. et al. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl Acad. Sci. USA 110, 15325–15329 (2013).
Martel, A. et al. Recent introduction of a chytrid fungus endangers western palearctic salamanders. Science 6209, 630–631 (2014).
Stegen, G. et al. Drivers of salamander extirpation mediated by Batrachochytrium salamandrivorans. Nature 544, 353–356 (2017).
McKenzie, V. J., Bowers, R. M., Fierer, N., Knight, R. & Lauber, C. L. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME. J. 6, 588–596 (2012).
Jani, A. J. & Briggs, C. J. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl Acad. Sci. USA 111, E5049–E5058 (2014).
Kueneman, J. G. et al. The amphibian skin-associated microbiome across species, space and life history stages. Mol. Ecol. 23, 1238–1250 (2014).
Bletz, M. C. et al. Host ecology rather than host phylogeny drives amphibian skin microbial community structure in the biodiversity hotspot of Madagascar. Front. Microbiol. 8, Article 1530 (2017).
Wolz, M. et al. Effects of host species and environment on the skin microbiome of Plethodontid salamanders. J. Anim. Ecol. 87, 341–353 (2017).
Longo, A. V., Savage, A. E., Hewson, I. & Zamudio, K. R. Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R. Soc. Open Sci. 2, 140377 (2015).
Sabino-Pinto, J. et al. Temporal changes in cutaneous bacterial communities of terrestrial- and aquatic-phase newts (Amphibia).Environ. Microbiol. 19, 3025–3038 (2017).
Bletz, M. C. et al. Amphibian skin microbiota exhibits temporal variation in community structure but stability of predicted Bd-inhibitory function. ISME. J. 11, 1521–1534 (2017).
Becker, M. H., Richards-Zawacki, C. L., Gratwicke, B. & Belden, L. K. The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki). Biol. Conserv. 176, 199–206 (2014).
Becker, C. G.., Longo, A. V., Haddad, C. F. B. & Zamudio, K. R. Land cover and forest connectivity alter the interactions among host, pathogen and skin microbiome.Proc. Biol. Sci. 284, pii: 20170582 (2017).
McKenzie, V. J. et al. The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690–704 (2017).
Agler, M. T. et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, 1–31 (2016).
Vavre, F. & Kremer, N. Microbial impacts on insect evolutionary diversification: from patterns to mechanisms. Curr. Opin. Insect Sci. 4, 29–34 (2014).
Webster, N. S. et al. Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6, 19324 (2016).
O’Brien, P. A., Morrow, K. M., Willis, B. & Bourne, D. Implications of ocean acidification for marine microorganisms from the free-living to the host-associated. Front. Mar. Sci 3, 47 (2016).
Longo, A. V. & Zamudio, K. R. Temperature variation, bacterial diversity and fungal infection dynamics in the amphibian skin. Mol. Ecol. 18, 4787–4797 (2017).
Longo, A. V. & Zamudio, K. R. Environmental fluctuations and host skin bacteria shift survival advantage between frogs and their fungal pathogen. ISME. J. 11, 349–361 (2017).
Novakova, E. et al. Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile virus. Front. Microbiol. 8, 526 (2017).
Corkrey, R. et al. The biokinetic spectrum for temperature. PLoS ONE. 11, 1–29 (2016).
Roller, B. R., Stoddard, S. F. & Schmidt, T. M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 18, 386–392 (2015).
Nemergut, D. R. et al. Decreases in average bacterial community rRNA operon copy number during succession. ISME. J. 10, 1147–1156 (2016).
Kerns, P. & Shade, A. Trait-based patterns of microbial dynamics in dormancy potential and heterotrophic strategy: case studies of resource-based and post-press succession.ISME J. 12, 2575–2581 (2018).
Czaran, T., Hoekstra, R. F. & Pagie, L. Chemical warfare between microbes. PNAS 99, 786–790 (2002).
Lennon, J. T. & Jones, S. E. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
Mittelbach, G. G. et al. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 (2007).
Wiens, J. J. Global patterns of diversification and species richness in amphibians. Am. Nat. 170, S86–S106 (2007).
Prest, T. L., Kimball, A. K., Kueneman, J. G. & McKenzie, V. J. Host‐associated bacterial community succession during amphibian development. Mol. Ecol. 27, 1992–2006 (2018).
Bahram, M. et al. Structure and function of the global topsoil microbiome. Nature 560, 233–237 (2018).
Staddon, W. J., Trevors, J. T., Duchesne, L. C. & Colombo, C. A. Soil microbial diversity and community structure across a climatic gradient in western Canada. Biodivers. Conserv. 7, 1081–1092 (1998).
Loudon, A. H. et al. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME. J. 8, 830–840 (2014).
Walke, J. B. et al. Amphibian skin may select for rare environmental microbes. ISME. J. 8, 2207–2217 (2014).
Belden, L. K. et al. Panamanian frog species host unique skin bacterial communities. Front. Microbiol. 6, 1171 (2015).
Sabino-Pinto, J. et al. Composition of the cutaneous bacterial community in Japanese amphibians: effects of captivity, host species, and body region.Microb. Ecol. 72, 460–469 (2016).
Rebollar, A. A. et al. Skin bacterial diversity of Panamanian frogs is associated with host susceptibility and presence of Batrachochytrium dendrobatidis.ISME J. 10, 1682–1695 (2016).
Bagley, S. T. Habitat association of Klebsiella species. Infect. Control. 6, 52–58 (1985).
Locey, K. J., Fisk, M. C. & Lennon, J. T. Microscale insight into microbial seed banks. Front. Microbiol. 7, 2040 (2017).
Stubbendieck, R. M., Vargas-Bautista, C. & Straight, P. D. Bacterial communities: interactions to scale. Front. Microbiol. 7, 1–19 (2016).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Amir, A. et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, 00191-16 (2017).
Cooper, N., Bielby, J., Thomas, G. H. & Purvis, A. Macroecology and extinction risk correlates of frogs. Glob. Ecol. Biogeogr. 17, 211–221 (2008).
Hedges, S. B., Dudley, J. & Kumar, S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006).
R Core Team, R: A Language and Environment for Statistical Computing (R Core Team, 2016).
Bates, D. M., Machler, M., Bolker, B. M. & Walker, S. C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2014).
Lüdecke, D. Data visualization for statistics in social science. R package https://doi.org/10.5281/zenodo.1308157 (2017).
Oksanen, J. et al. vegan: community ecology package. R package V.2, 4–2 (R Core Team, 2017).
Hothorn, T., Hornik, K., Van De Wiel, M. A. & Zeileis, A. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28, 1–23 (2008).
Acknowledgements
This study was supported by grants of National Science Foundation (DEB-1146284 to V.J.M.; IOS-1121758 to L.R.-S.; DEB-1310036 to A.V.L.), Templeton Foundation to V.J.M., Deutsche Forschungsgemeinschaft (DFG) to M.V. (VE247/9-1), CAPES to M.V. and C.F.B.H., FAPESP (#2013/50741-7) and CNPq to C.F.B.H., Simons Foundation (429440, WTW) to J.G.K., Deutscher Akademischer Austauschdienst (DAAD) to M.C.B., University of Costa Rica (Project 801-B2-029) and Costa Rican Ministry of Science and Technology (849-PINN-2015)-I to J.G.A., the Portuguese National Funds through FCT (Exploratory Research Project: IF/00209/2014/CP1256/CT0011) to A.C. and the National Research Foundation of Korea (2015R1D1A1A01057282) to B.W. We are indebted to M. Kondermann for her assistance with laboratory work, A. Borzee, R. Kakehashi and T. Kosch for their support in sample collection, and the Organization for Tropical Studies, La Selva Biological Station and G. Alvarado for field support in Costa Rica. All new sampling was done with the appropriate permits of national authorities, where required; in Costa Rica, of the Institutional Biodiversity Commission of the University of Costa Rica (Resolution 371 014) and the Costa Rican Ministry of Environment and Energy (Resolution 091-2012-SINAC).
Author information
Authors and Affiliations
Contributions
J.G.K., M.C.B., V.J.M., D.C.W. and M.V. conceived the study, coordinated the analyses and wrote the manuscript. J.G.K., M.C.B., D.C.W., G.B. and M.J. designed and performed data analysis. J.G.A., A.B., M.B., L.B., A.C., C.F.B.H., R.N.H., W.H., M.H., J.L.K., J.K., A.K., A.L., A.H.L., D.M., J.J.N., R.G.B.P., A.P.T., F.C.E.R., E.A.R., A.R., L.R.S., G.V.A., B.W., J.B.W., S.M.W., K.Z., I.Z.C. contributed materials and data. H.A., L.A., R.G. and M.J. performed laboratory work. P.J.K., R.S. and C.C.T. contributed to data analysis. All authors contributed to the development and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Results, Supplementary Figures 1–12, Supplementary Tables 1–17 and Supplementary References
Rights and permissions
About this article
Cite this article
Kueneman, J.G., Bletz, M.C., McKenzie, V.J. et al. Community richness of amphibian skin bacteria correlates with bioclimate at the global scale. Nat Ecol Evol 3, 381–389 (2019). https://doi.org/10.1038/s41559-019-0798-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0798-1
This article is cited by
-
Host phylogeny and environment shape the diversity of salamander skin bacterial communities
Animal Microbiome (2023)
-
The Mycobiome of Bats in the American Southwest Is Structured by Geography, Bat Species, and Behavior
Microbial Ecology (2023)
-
Factors Influencing Bacterial and Fungal Skin Communities of Montane Salamanders of Central Mexico
Microbial Ecology (2023)
-
From Alien Species to Alien Communities: Host- and Habitat-Associated Microbiomes in an Alien Amphibian
Microbial Ecology (2023)
-
Environmental and Anthropogenic Factors Shape the Skin Bacterial Communities of a Semi-Arid Amphibian Species
Microbial Ecology (2023)