Abstract
Diatoms are one of the most abundant and diverse groups of phytoplankton and play a major role in marine ecosystems and the Earth’s biogeochemical cycles. Here we combine DNA metabarcoding data from the Tara Oceans expedition with palaeoenvironmental data and phylogenetic models of diversification to analyse the diversity dynamics of marine diatoms. We reveal a primary effect of variation in carbon dioxide partial pressure (pCO2) on early diatom diversification, followed by a major burst of diversification in the late Eocene epoch, after which diversification is chiefly affected by sea level, an influx of silica availability and competition with other planktonic groups. Our results demonstrate a remarkable heterogeneity of diversification dynamics across diatoms and suggest that a changing climate will favour some clades at the expense of others.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are included as Supplementary Data, or EBI accession numbers are provided.
Change history
13 November 2018
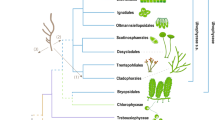
In the version of this Article originally published, the authors did not give credit to David G. Mann for the four microscopic images used in Fig. 1a. This has now been amended in all versions of the Article.
References
Katz, M. E., Finkel, Z. V., Grzebyk, D., Knoll, A. H. & Falkowski, P. G. Evolutionary trajectories and biogeochemical impacts of marine eukaryotic phytoplankton. Annu. Rev. Ecol. Evol. Syst. 35, 523–556 (2004).
Kooistra, W. H., Gersonde, R., Medlin, L. K. & Mann, D. G. in Evolution of Primary Producers in the Sea (eds Falkowski, P. G. & Knoll, A. H.) 207–249 (Academic, Burlington, 2007).
Katz, M. E. et al. Stepwise transition from the Eocene greenhouse to the Oligocene icehouse. Nat. Geosci. 1, 329–334 (2008).
Baatsen, M., von der Heydt, A., Kliphuis, M., Viebahn, J. & Dijkstra, H. Multiple states in the late Eocene ocean circulation. Glob. Planet. Change 163, 18–28 (2018).
Livermore, R., Hillenbrand, C.-D. & Meredith, M. Drake Passage and Cenozoic climate: an open and shut case?. Geochem. Geophys. Geosyst. 8, Q01005 (2007).
Bijl, P. K. et al. Eocene cooling linked to early flow across the Tasmanian gateway. Proc. Natl Acad. Sci. USA 110, 9645–9650 (2013).
Carter, A., Riley, T. R., Hillenbrand, C.-D. & Rittner, M. Widespread Antarctic glaciation during the late Eocene. Earth Planet. Sci. Lett. 458, 49–57 (2017).
Miller, K. G., Wright, J. & Fairbanks, R. G. Unlocking the ice house: Oligocene–Miocene oxygen isotopes, eustasy, and margin erosion. J. Geophys. Res. 96, 6829–6848 (1991).
Cermeño, P., Falkowski, P. G., Romero, O. E., Schaller, M. F. & Vallina, S. M. Continental erosion and the Cenozoic rise of marine diatoms. Proc. Natl Acad. Sci. USA 112, 4239–4244 (2015).
Spencer-Cervato, C. The Cenozoic deep sea microfossil record: explorations of the DSDP/ODP sample set using the Neptune database. Palaeontol. Electronica 2, 270 (1999).
Lazarus, D., Barron, J., Renaudie, J., Diver, P. & Türke, A. Cenozoic planktonic marine diatom diversity and correlation to climate change. PLoS One 9, e84857 (2014).
Barker, P. F. Scotia Sea regional tectonic evolution: implications for mantle flow and palaeocirculation. Earth Sci. Rev. 55, 1–39 (2001).
Scher, H. D. & Martin, E. E. Timing and climatic consequences of the opening of Drake Passage. Science 312, 428–430 (2006).
Falkowski, P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360 (2004).
Finkel, Z. V., Katz, M. E., Wright, J. D., Schofield, O. M. E. & Falkowski, P. G. Climatically driven macroevolutionary patterns in the size of marine diatoms over the Cenozoic. Proc. Natl Acad. Sci. USA 102, 8927–8932 (2005).
Kooistra, W. H. et al. Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist 159, 177–193 (2008).
Ezard, T. H. G., Aze, T., Pearson, P. N. & Purvis, A. Interplay between changing climate and species’ ecology drives macroevolutionary dynamics. Science 332, 349–351 (2011).
Yasuhara, M., Tittensor, D. P., Hillebrand, H. & Worm, B. Combining marine macroecology and palaeoecology in understanding biodiversity: microfossils as a model. Biol. Rev. 92, 199–215 (2017).
de Vargas, C. et al. Oceanic plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
Malviya, S. et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl Acad. Sci. USA 113, E1516–E1525 (2016).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013).
Quince, C., Curtis, T. P. & Sloan, W. T. The rational exploration of microbial diversity. ISME J. 2, 997–1006 (2008).
Stadler, T. Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl Acad. Sci. USA 108, 6187–6192 (2011).
Harwood, D. M. in Geology and Paleontology of Seymour Island Antarctic Peninsula (eds Feldmann, R. M. & Woodburne, M. O.) 55–130 (Geological Society of America Memoirs Vol. 169, Geological Society of America, 1988).
Jordan, R. & Stickley, C. E. The Diatoms: Applications for the Environmental and Earth Sciences (eds Smol, J. P. & Stoermer, E. F.) 424–453 (Cambridge Univ. Press, Cambridge, 2010).
Rabosky, D. L. & Sorhannus, U. Diversity dynamics of marine planktonic diatoms across the Cenozoic. Nature 457, 183–186 (2009).
Morlon, H., Parsons, T. L. & Plotkin, J. B. Reconciling molecular phylogenies with the fossil record. Proc. Natl Acad. Sci. USA 108, 16327–16332 (2011).
Condamine, F. L., Rolland, J. & Morlon, H. Macroevolutionary perspectives to environmental change. Ecol. Lett. 16(Suppl. 1), 72–85 (2013).
Lewitus, E. & Morlon, H. Detecting environment-dependent diversification from phylogenies: a simulation study and some empirical illustrations. Syst. Biol. 67, 576–593 (2018).
Falkowski, P. G., Fenchel, T. & Delong, E. F. The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039 (2008).
Phillimore, A. B. & Price, T. D. Density-dependent cladogenesis in birds. PLoS Biol. 6, e71 (2008).
McPeek, M. A. The ecological dynamics of clade diversification and community assembly. Am. Nat. 172, E270–E284 (2008).
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015).
Lazarus, D. B., Kotrc, B., Wulf, G. & Schmidt, D. N. Radiolarians decreased silicification as an evolutionary response to reduced Cenozoic ocean silica availability. Proc. Natl Acad. Sci. USA 106, 9333–9338 (2009).
Elderfield, H. et al. Evolution of ocean temperature and ice volume through the mid-Pleistocene climate transition. Science 337, 704–709 (2012).
Zachos, J. C., Dickens, G. R. & Zeebe, R. E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451, 279–283 (2008).
Vallina, S. M. et al. Global relationship between phytoplankton diversity and productivity in the ocean. Nat. Commun. 5, 4299 (2014).
Armbrust, E. V. The life of diatoms in the world’s oceans. Nature 459, 185–192 (2009).
Dorrell, R. G. & Smith, A. G. Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell 10, 856–868 (2011).
Gooday, A. J. Biological responses to seasonally varying fluxes of organic matter to the ocean floor: a review. J. Oceanogr. 58, 305–332 (2002).
Barnosky, A. D. Distinguishing the effects of the Red Queen and Court Jester on Miocene mammal evolution in the northern Rocky Mountains. J. Vert. Paleontol. 21, 172–185 (2001).
Van Valen, L. A new evolutionary law. Evol. Theory 1, 1–30 (1973).
Benton, M. J. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732 (2009).
Medlin, L. K. & Kaczmarska, I. Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 43, 245–270 (2004).
Sims, P. A., Mann, D. G. & Medlin, L. K. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402 (2006).
Medlin, L. K., Sato, S., Mann, D. G. & Kooistra, W. H. C. F. Molecular evidence confirms sister relationship of Ardissonea, Climacosphenia, and Toxarium within the bipolar centric diatoms (Bacillariophyta, Mediophyceae), and cladistic analyses confirm that extremely elongated shape has arisen twice in the diatoms. J. Phycol. 44, 1340–1348 (2008).
Williams, D. M. & Kociolek, J. P. Pursuit of a natural classification of diatoms: history, monophyly and the rejection of paraphyletic taxa. Eur. J. Phycol. 42, 313–319 (2007).
Theriot, E. C., Cannone, J. J., Gutell, R. R. & Alverson, A. J. The limits of nuclear encoded SSU rDNA for resolving the diatom phylogeny. Eur. J. Phycol. 44, 277–290 (2009).
Rabosky, D. L. Challenges in the estimation of extinction from molecular phylogenies: a response to Beaulieu and O’Meara: brief communication. Evolution 70, 218–228 (2016).
Hunt, G. & Slater, G. Integrating paleontological and phylogenetic approaches to macroevolution. Annu. Rev. Ecol. Evol. Syst. 47, 189–213 (2016).
DeConto, R. M. & Pollard, D. Rapid Cenozoic glaciation of Antarctica induced by declining atmospheric CO2. Nature 421, 245–249 (2003).
Tripati, A. & Darby, D. Evidence for ephemeral middle Eocene to early Oligocene Greenland glacial ice and pan-Arctic sea ice. Nat. Commun. 9, 1038 (2018).
Miller, K. G. The Phanerozoic record of global sea-level change. Science 310, 1293–1298 (2005).
Richter, F. M., Rowley, D. B. & DePaolo, D. J. Sr isotope evolution of seawater: the role of tectonics. Earth Planet. Sci. Lett. 109, 11–23 (1992).
Misra, S. & Froelich, P. N. Lithium isotope history of Cenozoic seawater: changes in silicate weathering and reverse weathering. Science 335, 818–823 (2012).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Larkin, M. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723 (1974).
Liu, K., Linder, C. R. & Warnow, T. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PloS One 6, e27731 (2011).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS One 5, e9490 (2010).
Pesant, S. et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2, 150023 (2015).
De Vargas, C., Audic, S., Tara Oceans Consortium Coordinators & Tara Oceans Expedition Participants Total V9 rDNA Information Organized at the OTU Level for the Tara Oceans Expedition (2009–2012) (Pangaea, 2017); https://doi.org/10.1594/PANGAEA.873275
Bork, P. et al. Tara Oceans studies plankton at planetary scale. Science 348, 873–873 (2015).
Edgar, R. C. Search and clustering orders of magnitude faster than blast. Bioinformatics 26, 2460–2461 (2010).
Britton, T., Anderson, C. L., Jacquet, D., Lundqvist, S. & Bremer, K. Estimating divergence times in large phylogenetic trees. Syst. Biol. 56, 741–752 (2007).
Kooistra, W. H. & Medlin, L. K. Evolution of the diatoms (Bacillariophyta). IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Mol. Phylogenet. Evol. 6, 391–407 (1996).
Sorhannus, U. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar. Micropaleontol. 65, 1–12 (2007).
Paradis, E., Claude, J. & Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290 (2004).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Lewitus, E. & Morlon, H. Characterizing and comparing phylogenies from their Laplacian spectrum. Syst. Biol. 65, 495–507 (2016).
Morlon, H. et al. Spatial patterns of phylogenetic diversity. Ecol. Lett. 14, 141–149 (2011).
Sichel, H. S. On a distribution representing sentence-length in written prose. J. R. Stat. Soc. Series A 137, 25–34 (1974).
Curtis, T. P., Sloan, W. T. & Scannell, J. W. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA 99, 10494–10499 (2002).
Hong, S.-H., Bunge, J., Jeon, S.-O. & Epstein, S. S. Predicting microbial species richness. Proc. Natl Acad. Sci. USA 103, 117–122 (2006).
Spiegelhalter, D. J., Best, N. G., Carlin, B. P. & van der Linde, A. Bayesian measures of model complexity and fit. J. R. Stat. Soc. Series B 64, 583–639 (2002).
Revell, L. J. & Reynolds, R. G. A new Bayesian method for fitting evolutionary models to comparative data with intraspecific variation. Evolution 66, 2697–2707 (2012).
Morlon, H. et al. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol. 7, 589–597 (2016).
Royer, D. L., Berner, R. A., Montañez, I. P., Tabor, N. J. & Beerling, D. J. CO2 as a primary driver of phanerozoic climate. GSA Today 14, 4–10 (2004).
Mayhew, P. J., Jenkins, G. B. & Benton, T. G. A long-term association between global temperature and biodiversity, origination and extinction in the fossil record. Proc. Biol. Sci. 275, 47–53 (2008).
Hannisdal, B. & Peters, S. E. Phanerozoic Earth system evolution and marine biodiversity. Science 334, 1121–1124 (2011).
Lazarus, D. Neptune: a marine micropaleontology database. Math. Geol. 26, 817–832 (1994).
Alroy, J. Geographical, environmental and intrinsic biotic controls on Phanerozoic marine diversification. Palaeontology 53, 1211–1235 (2010).
Hansen, J., Sato, M., Russell, G. & Kharecha, P. Climate sensitivity, sea level and atmospheric carbon dioxide. Phil. Trans. R. Soc. A 371, 20120294 (2013).
Nee, S., Mooers, A. O. & Harvey, P. H. Tempo and mode of evolution revealed from molecular phylogenies. Proc. Natl Acad. Sci. USA 89, 8322–8326 (1992).
der Voo, R. V., Spakman, W. & Bijwaard, H. Tethyan subducted slabs under India. Earth Planet. Sci. Lett. 171, 7–20 (1999).
Schulte, P. et al. The Chicxulub asteroid impact and mass extinction at the Cretaceous–Paleogene boundary. Science 327, 1214–1218 (2010).
Kennett, J. P. & Stott, L. D. Abrupt deep-sea warming, palaeoceanographic changes and benthic extinctions at the end of the Palaeocene. Nature 353, 225–229 (1991).
Yasuhara, M., Hunt, G. & Okahashi, H. Quaternary deep-sea ostracods from the north-western Pacific Ocean: global biogeography and Drake-Passage, Tethyan, Central American and Arctic pathways. J. Syst. Palaeontol. https://doi.org/10.1080/14772019.2017.1393019 (2017).
Acknowledgements
We thank J. Clavel, G. Sommeria-Klein, O. Maliet, M. Manceau and O. Missa for comments on the manuscript. E.L. thanks E. Charles for discussion. Funding was provided through a European Research Council Consolidator grant (ERC-CoG-PANDA) attributed to H.M. C.B. acknowledges funding from the ERC Advanced Award Diatomite (294823), the LouisD Foundation and the French Government ‘Investissements d’Avenir’ programmes MEMO LIFE (ANR-10-LABX-54), PSL* Research University (ANR-1253 11-IDEX-0001-02) and OCEANOMICS (ANR-11-BTBR-0008). This Article is contribution no. 80 of the Tara Oceans project.
Author information
Authors and Affiliations
Contributions
E.L., H.M. and C.B. conceived the study. E.L. analysed the data. L.B., S.M. and C.B. contributed data. E.L. and H.M. wrote the manuscript. C.B. contributed substantially to revisions. The views expressed are those of the authors and should not be construed to represent the positions of the US Army or the Department of Defense.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary tables and figures
Supplementary Data 1
V9-18S ribosomal DNA sequences for 22,0018 global samples
Supplementary Data 2
Backbone phylogenies constructed using the Protist Ribosomal Database using two multiple alignment methods (MAFFT, CLUSTALW) and two tree reconstruction methods (FastTree, RaXML)
Supplementary Data 3
Consensus phylogenies for 19,197 diatom OTUs constructed from four backbone phylogenies
Supplementary Data 4
Uclust output of V9-18S ribosomal DNA sequences for 22,0018 global samples defining diatom OTUs
Supplementary Data 5
Taxonomic assignment of V9-18S ribosomal DNA sequences from global samples
Rights and permissions
About this article
Cite this article
Lewitus, E., Bittner, L., Malviya, S. et al. Clade-specific diversification dynamics of marine diatoms since the Jurassic. Nat Ecol Evol 2, 1715–1723 (2018). https://doi.org/10.1038/s41559-018-0691-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-018-0691-3
This article is cited by
-
Decoupling speciation and extinction reveals both abiotic and biotic drivers shaped 250 million years of diversity in crocodile-line archosaurs
Nature Ecology & Evolution (2023)
-
Comparative analysis of full-length mitochondrial genomes of five Skeletonema species reveals conserved genome organization and recent speciation
BMC Genomics (2021)
-
Phytoplankton biodiversity and the inverted paradox
ISME Communications (2021)
-
Global radiation in a rare biosphere soil diatom
Nature Communications (2020)