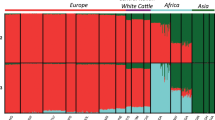
Abstract
Species of the Bos genus, including taurine cattle, zebu, gayal, gaur, banteng, yak, wisent and bison, have been domesticated at least four times and have been an important source of meat, milk and power for many human cultures. We sequence the genomes of gayal, gaur, banteng, wisent and bison, and provide population genomic sequencing of an additional 98 individuals. We use these data to determine the phylogeny and evolutionary history of these species and show that the threatened gayal is an independent species or subspecies. We show that there has been pronounced introgression among different members of this genus, and that it in many cases has involved genes of considerable adaptive importance. For example, genes under domestication selection in cattle (for example, MITF) were introgressed from domestic cattle to yak. Also, genes in the response-to-hypoxia pathway (for example, EGLN1, EGLN2 and HIF3a) have been introgressed from yak to Tibetan cattle, probably facilitating their adaptation to high altitude. We also validate that there is an association between the introgressed EGLN1 allele and haemoglobin and red blood cell concentration. Our results illustrate the importance of introgression as a source of adaptive variation and during domestication, and suggest that the Bos genus evolves as a complex of genetically interconnected species with shared evolutionary trajectories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Soubrier, J. et al. Early cave art and ancient DNA record the origin of European bison. Nat. Commun. 7, 13158 (2014).
Gautier, M. et al. Deciphering the wisent demographic and adaptive histories from individual whole-genome sequences. Mol. Biol. Evol. 33, 2801–2814 (2016).
Wang, K. et al. The genome sequence of the wisent (Bison bonasus). GigaScience 6, 1–5 (2017).
Qiu, Q. et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 6, 10283 (2015).
Medugorac, I. et al. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nat. Genet. 49, 470–475 (2017).
Daetwyler, H. D. et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46, 858–865 (2014).
Qiu, Q. et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949 (2012).
Newman, J. H. et al. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat. Commun. 6, 6863 (2015).
Decker, J. E. et al. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc. Natl Acad. Sci. USA 106, 18644–18649 (2009).
Hassanin, A., An, J., Ropiquet, A., Nguyen, T. T. & Couloux, A. Combining multiple autosomal introns for studying shallow phylogeny and taxonomy of Laurasiatherian mammals: application to the tribe Bovini (Cetartiodactyla, Bovidae). Mol. Phylogenet. Evol. 66, 766–775 (2013).
Buntjer, J. B., Otsen, M., Nijman, I. J., Kuiper, M. T. R. & Lenstra, J. A. Phylogeny of bovine species based on AFLP fingerprinting. Heredity 88, 46–51 (2002).
Ma, G. et al. Phylogenetic relationships and status quo of colonies for gayal based on analysis of cytochrome b gene partial sequences. J. Genet. Genom. 34, 413–419 (2007).
Baig, M. et al. Mitochondrial DNA diversity and origin of Bos frontalis. Curr. Sci. 104, 115–120 (2013).
Gou, X., Wang, Y., Yang, S., Deng, W. & Mao, H. Genetic diversity and origin of gayal and cattle in Yunnan revealed by mtDNA control region and SRY gene sequence variation. J. Anim. Breed. Genet. 127, 154–160 (2010).
Mei, C. et al. Whole-genome sequencing of the endangered bovine species gayal (Bos frontalis) provides new insights into its genetic features. Sci. Rep. 6, 19787 (2016).
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011).
Nielsen, R. et al. Tracing the peopling of the world through genomics. Nature 541, 302–310 (2017).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Mirarab, S. et al. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548 (2014).
Liu, L., Yu, L. & Edwards, S. V. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Biol. 10, 302 (2010).
Verkaar, E. L. C., Nijman, I. J., Beeke, M., Hanekamp, E. & Lenstra, J. A. Maternal and paternal lineages in cross-breeding bovine species. Has wisent a hybrid origin?. Mol. Biol. Evol 21, 1165–1170 (2004).
Hobolth, A., Christensen, O. F., Mailund, T. & Schierup, M. H. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3, e7 (2007).
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2014).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012).
Wecek, K. et al. Complex admixture preceded and followed the extinction of wisent in the wild. Mol. Biol. Evol. 34, 598–612 (2017).
Durand, E. Y., Patterson, N., Reich, D. & Slatkin, M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011).
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010).
Bosse, M. et al. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat. Commun. 5, 4392 (2014).
Hofer, E., Sobanov, Y., Brostjan, C., Lehrach, H. & Düchler, M. The centromeric part of the human natural killer (NK) receptor complex: lectin-like receptor genes expressed in NK, dendritic and endothelial cells. Immunol. Rev. 181, 5–19 (2001).
Kao, H.-T. et al. A third member of the synapsin gene family. Proc. Natl Acad. Sci. USA 95, 4667–4672 (1998).
Porton, B. et al. Mice lacking synapsin III show abnormalities in explicit memory and conditioned fear. Genes Brain Behav. 9, 257–268 (2009).
Per, J. Behavior genetics and the domestication of animals. Annu. Rev. Anim. Biosci. 2, 85–104 (2014).
Driscoll, C. A., Macdonald, D. W. & O’Brien, S. J. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl Acad. Sci. USA 106, 9971–9978 (2009).
Suzuki, G. et al. Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum. Mol. Genet. 18, 1652–1660 (2009).
Carneiro, M. et al. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 10, e1003519 (2014).
Li, Y. et al. Artificial selection on brain-expressed genes during the domestication of dog. Mol. Biol. Evol. 30, 1867–1876 (2013).
Wang, G.-d. et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 4, 1860 (2013).
The-Bovine-HapMap-Consortium Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532 (2009).
Maples, B. K., Gravel, S., Kenny, E. E. & Bustamante, C. D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013).
Huerta-Sánchez, E. et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014).
Lorenzo, F. R. et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 46, 951–956 (2014).
Bigham, A. W. & Lee, F. S. Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 28, 2189–2204 (2014).
Storz, J. F., Scott, G. R. & Cheviron, Z. A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 213, 4125–4136 (2010).
Chen, F. H. et al. Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P. Science 347, 248–250 (2015).
Miao, B., Wang, Z. & Li, Y. Genomic analysis reveals hypoxia adaptation in the Tibetan mastiff by introgression of the gray wolf from the Tibetan Plateau. Mol. Biol. Evol. 34, 734–743 (2017).
Hsiao, J. J. & Fisher, D. E. The roles of microphthalmia transcription factor and pigmentation in melanoma. Arch. Biochem. Biophys. 563, 28–34 (2015).
Kim, J. et al. The genome landscape of indigenous African cattle. Genome Biol. 18, 34 (2017).
Qanbari, S. et al. Classic selective sweeps revealed by massive sequencing in cattle. PLoS Genet. 10, e1004148 (2012).
Hayes, B. J., Pryce, J., Chamberlain, A. J., Bowman, P. J. & Goddard, M. E. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6, e1001139 (2010).
Kong, Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153 (2011).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Popescu, A.-A., Huber, K. T. & Paradis, E. ape 3.0: new tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics 28, 1536–1537 (2012).
Mailund, T. et al. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet. 8, e1003125 (2012).
Mailund, T., Dutheil, J. Y., Hobolth, A., Lunter, G. & Schierup, M. H. Estimating divergence time and ancestral effective population size of Bornean and Sumatran orangutan subspecies using a coalescent hidden Markov model. PLoS Genet. 7, e1001319 (2011).
Kumar, S. & Subramanian, S. Mutation rates in mammalian genomes. Proc. Natl Acad. Sci. USA 99, 803–808 (2002).
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013).
Maples, B. K., Gravel, S., Kenny, E. E. & Bustamante, C. D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Reimand, J. et al. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44, W83–W89 (2016).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2008).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No XDB13020600), National Natural Science Foundation of China (91731304, 31321002, 31272418, 31561143010), the Chinese 973 program (2013CB835204), Animal Branch of the Germplasm Bank of Wild Species (GBOWS) and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R621). D.-D.W. was supported by the Youth Innovation Promotion Association, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
Y.-P.Z., D.-D.W., R.N. and Q.Z. lead the project, and designed and conceived the study. D.-D.W., S.W., X.-D.D. and R.N. prepared the manuscript. D.-D.W., S.W., X.-D.D., Y. Z., Y.L. and M.-S.W. performed the data analysis. J.M.W., M.T. and O.F. performed some sampling and experiments. All authors read the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures and tables 1–4
Supplementary table 5
Genetic introgression between gayal and zebu
Supplementary table 6
Genetic introgression between bali cattle and zebu
Supplementary table 7
Gene enrichment analysis genes within regions showing genetic introgression between zebu and gayal
Supplementary table 8
Gene enrichment analysis genes within regions showing genetic introgression between zebu and bali cattle
Supplementary table 9
Genetic introgression between yak and Tibetan cattle
Supplementary table 10
Gene enrichment analysis genes within regions showing genetic introgression between yak and Tibetan cattle
Rights and permissions
About this article
Cite this article
Wu, DD., Ding, XD., Wang, S. et al. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol 2, 1139–1145 (2018). https://doi.org/10.1038/s41559-018-0562-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-018-0562-y
This article is cited by
-
Insight into the evolutionary and domesticated history of the most widely cultivated mushroom Agaricus bisporus via mitogenome sequences of 361 global strains
BMC Genomics (2023)
-
Evolutionary origin of genomic structural variations in domestic yaks
Nature Communications (2023)
-
Global dispersal and adaptive evolution of domestic cattle: a genomic perspective
Stress Biology (2023)
-
Structural variation and introgression from wild populations in East Asian cattle genomes confer adaptation to local environment
Genome Biology (2023)
-
Global genetic diversity, introgression, and evolutionary adaptation of indicine cattle revealed by whole genome sequencing
Nature Communications (2023)