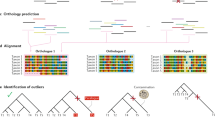
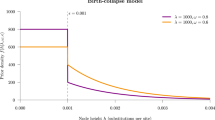
Abstract
Bayesian methods have become very popular in molecular phylogenetics due to the availability of user-friendly software for running sophisticated models of evolution. However, Bayesian phylogenetic models are complex, and analyses are often carried out using default settings, which may not be appropriate. Here we summarize the major features of Bayesian phylogenetic inference and discuss Bayesian computation using Markov chain Monte Carlo (MCMC) sampling, the diagnosis of an MCMC run, and ways of summarizing the MCMC sample. We discuss the specification of the prior, the choice of the substitution model and partitioning of the data. Finally, we provide a list of common Bayesian phylogenetic software packages and recommend appropriate applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rannala, B. & Yang, Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311 (1996).
Mau, B. & Newton, M. A. Phylogenetic inference for binary data on dendograms using Markov chain Monte Carlo. J. Comp. Graph. Stat. 6, 122–131 (1997).
Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294, 2310–2314 (2001).
Wilfert, L. et al. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 351, 594–597 (2016).
Pybus, O. G. et al. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc. Natl Acad. Sci. USA 109, 15066–15071 (2012).
Faria, N. R. et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61 (2014).
Lemey, P., Rambaut, A., Welch, J. J. & Suchard, M. A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010).
Bloomquist, E. W., Lemey, P. & Suchard, M. A. Three roads diverged? Routes to phylogeographic inference. Trends Ecol. Evol. 25, 626–632 (2010).
Nascimento, F. F. et al. The role of historical barriers in the diversification processes in open vegetation formations during the Miocene/Pliocene using an ancient rodent lineage as a model. PLoS ONE 8, e61924 (2013).
Werneck, F. P., Leite, R. N., Geurgas, S. R. & Rodrigues, M. T. Biogeographic history and cryptic diversity of saxicolous Tropiduridae lizards endemic to the semiarid Caatinga. BMC Evol. Biol. 15, 94 (2015).
Merckx, V. S. F. T. et al. Evolution of endemism on a young tropical mountain. Nature 524, 347–350 (2015).
Hoorn, C. et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330, 927–931 (2010).
Prum, R. O. et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526, 569–573 (2015).
dos Reis, M. et al. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 25, 2939–2950 (2015).
Meredith, R. W. et al. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (2011).
Nascimento, F. F. et al. Evolution of endogenous retroviruses in the Suidae: evidence for different viral subpopulations in African and Eurasian host species. BMC Evol. Biol. 11, 139 (2011).
Jarvis, E. D. et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331 (2014).
Misof, B. et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014).
Raymann, K., Brochier-Armanet, C. & Gribaldo, S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl Acad. Sci. USA 112, 6670–6675 (2015).
Foley, N. M., Springer, M. S. & Teeling, E. C. Mammal madness: is the mammal tree of life not yet resolved? Phil. Trans. R. Soc. B 371, 20150140 (2016).
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 (1953).
Hastings, W. K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970).
Liu, L., Xi, Z., Wu, S., Davis, C. C. & Edwards, S. V. Estimating phylogenetic trees from genome-scale data. Ann. NY Acad. Sci. 1360, 36–53 (2015).
Xu, B. & Yang, Z. Challenges in species tree estimation under the multispecies coalescent model. Genetics 204, 1353–1368 (2016).
Szöllosi, G. J., Tannier, E., Daubin, V. & Boussau, B. The inference of gene trees with species trees. Syst. Biol. 64, e42–e62 (2015).
Yang, Z. Molecular Evolution: A Statistical Approach (Oxford Univ. Press, Oxford, 2014).
Lewis, P. O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925 (2001).
Redelings, B. D. & Suchard, M. A. Joint Bayesian estimation of alignment and phylogeny. Syst. Biol. 54, 401–418 (2005).
Löytynoja, A. & Goldman, N. Uniting alignments and trees. Science 324, 1528–1529 (2009).
Chatzou, M. et al. Multiple sequence alignment modeling: methods and applications. Brief. Bioinform. 17, 1009–1023 (2016).
Altenhoff, A. M. & Dessimoz, C. Inferring orthology and paralogy. Methods Mol. Biol. 855, 259–279 (2012).
Altenhoff, A. M. et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 43, D240–D249 (2015).
Dimmic, M. in Statistical Methods in Molecular Evolution (ed. Nielsen, R.) 259–287 (Springer, New York, 2005).
Liò, P. & Goldman, N. Models of molecular evolution and phylogeny. Genome Res. 8, 1233–1244 (1998).
Jukes, T. H. & Cantor, C. R. in Mammalian Protein Metabolism (ed. Munro, H. N.) 21–132 (Academic, New York, 1969).
Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17, 57–86 (1986).
Yang, Z. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 39, 105–111 (1994).
Zharkikh, A. Estimation of evolutionary distances between nucleotide sequences. J. Mol. Evol. 39, 315–329 (1994).
Mayrose, I., Graur, D., Ben-Tal, N. & Pupko, T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 21, 1781–1791 (2004).
Yang, Z., Lauder, I. J. & Lin, H. J. Molecular evolution of the hepatitis B virus genome. J. Mol. Evol. 41, 587–596 (1995).
Yang, Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11, 367–372 (1996).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Keane, T. M., Creevey, C. J., Pentony, M. M., Naughton, T. J. & McInerney, J. O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6, 29 (2006).
Lanfear, R., Calcott, B., Ho, S. Y. & Guindon, S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701 (2012).
Hasegawa, M., Kishino, H. & Yano, T. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985).
Hoff, M., Orf, S., Riehm, B., Darriba, D. & Stamatakis, A. Does the choice of nucleotide substitution models matter topologically? BMC Bioinform. 17, 143 (2016).
Huelsenbeck, J. & Rannala, B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst. Biol. 53, 904–913 (2004).
Wright, A. M., Lloyd, G. T. & Hillis, D. M. Modeling character change heterogeneity in phylogenetic analyses of morphology through the use of priors. Syst. Biol. 65, 602–611 (2016).
Felsenstein, J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25, 471–492 (1973).
Felsenstein, J. Inferring Phylogenies (Sinauer Associates, Sunderland, 2004).
Ronquist, F. et al. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999 (2012).
Heath, T. A., Huelsenbeck, J. P. & Stadler, T. The fossilized birth-death process for coherent calibration of divergence-time estimates. Proc. Natl Acad. Sci. USA 111, E2957–E2966 (2014).
O’Reilly, J. E., dos Reis, M. & Donoghue, P. C. Dating tips for divergence-time estimation. Trends Genet. 31, 637–650 (2015).
Rannala, B. Identifiability of parameters in MCMC Bayesian inference of phylogeny. Syst. Biol. 51, 754–760 (2002).
Gu, X., Fu, Y. X. & Li, W. H. Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites. Mol. Biol. Evol. 12, 546–557 (1995).
Sullivan, J., Swofford, D. L. & Naylor, G. J. The effect of taxon sampling on estimating rate heterogeneity parameters of maximum-likelihood models. Mol. Biol. Evol. 16, 1347–1356 (1999).
Yang, Z. The BPP program for species tree estimation and species delimitation. Curr. Zool. 61, 854–865 (2015).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Shapiro, B., Rambaut, A. & Drummond, A. J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23, 7–9 (2006).
Yang, Z. & Rannala, B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226 (2006).
Nylander, J. A., Ronquist, F., Huelsenbeck, J. P. & Nieves-Aldrey, J. L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67 (2004).
Maddison, W. P. Gene trees in species trees. Syst. Biol. 46, 523–536 (1997).
Nichols, R. Gene trees and species tree are not the same. Trends Ecol. Evol. 16, 358–364 (2001).
Liu, L. & Pearl, D. K. Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst. Biol. 56, 504–514 (2007).
Edwards, S. V. et al. Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol. Phylogenet. Evol. 94, 447–462 (2016).
Vijaykrishna, D., Mukerji, R. & Smith, G. J. D. RNA virus reassortment: an evolutionary mechanism for host jumps and immune evasion. PLoS Pathog. 11, e1004902 (2015).
Ronquist, F., van der Mark, P. & Huelsenbeck, J. P. in The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing (eds Lemey, P. et al.) 210–236 (Cambridge Univ. Press, New York, 2009).
Brown, J. M., Hedtke, S. M., Lemmon, A. R. & Lemmon, E. M. When trees grow too long: investigating the causes of highly inaccurate bayesian branch-length estimates. Syst. Biol 59, 145–161 (2010).
Rannala, B., Zhu, T. & Yang, Z. Tail paradox, partial identifiability, and influential priors in Bayesian branch length inference. Mol. Biol. Evol. 29, 325–335 (2012).
dos Reis, M., Zhu, T. & Yang, Z. The impact of the rate prior on Bayesian estimation of divergence times with multiple loci. Syst. Biol. 63, 555–565 (2014).
Drummond, A. J., Ho, S. Y., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006).
Yang, Z. & Rannala, B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 14, 717–724 (1997).
Rannala, B. & Yang, Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164, 1645–1656 (2003).
Ho, S. Y. & Phillips, M. J. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380 (2009).
Thorne, J. L., Kishino, H. & Painter, I. S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 15, 1647–1657 (1998).
Rannala, B. & Yang, Z. Inferring speciation times under an episodic molecular clock. Syst. Biol. 56, 453–466 (2007).
dos Reis, M., Donoghue, P. C. & Yang, Z. Bayesian molecular clock dating of species divergences in the genomics era. Nat. Rev. Genet. 17, 71–80 (2016).
Yang, Z. & Rodriguez, C. E. Searching for efficient Markov chain Monte Carlo proposal kernels. Proc. Natl Acad. Sci. USA 110, 19307–19312 (2013).
Green, P. J. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732 (1995).
Lakner, C., van der Mark, P., Huelsenbeck, J. P., Larget, B. & Ronquist, F. Efficiency of Markov chain Monte Carlo tree proposals in Bayesian phylogenetics. Syst. Biol. 57, 86–103 (2008).
Green, P. J. & Han, X. L. in Stochastic Models, Statistical Methods, and Algorithms in Image Analysis (eds Barone, P. et al.) 142–164 (Springer, New York, 1992).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2017).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v.1.6 (2014); http://beast.community/tracer.
Solís-Lemus, C., Knowles, L. L. & Ané, C. Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 69, 492–507 (2015).
Chen, M.-H., Kuo, L. & Lewis, P. Bayesian Phylogenetics: Methods, Algorithms, and Applications (Chapman & Hall/CRC, Boca Raton,2014).
Gelman, A. et al. Bayesian Data Analysis (Chapman & Hall/CRC, Boca Raton, 2013).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Höhna, S. et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65, 726–736 (2016).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Lewis, P. O., Holder, M. T. & Swofford, D. L. Phycas: software for Bayesian phylogenetic analysis. Syst. Biol. 64, 525–531 (2015).
Lewis, P. O., Holder, M. T. & Holsinger, K. E. Polytomies and Bayesian phylogenetic inference. Syst. Biol. 54, 241–253 (2005).
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009).
Beerli, P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345 (2006).
Hey, J. & Nielsen, R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl Acad. Sci. USA 104, 2785–2790 (2007).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Rabosky, D. L. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 (2014).
Nylander, J. A., Wilgenbusch, J. C., Warren, D. L. & Swofford, D. L. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583 (2008).
Efron, B. & Tibshirani, R. J. An Introduction to the Bootstrap (Chapman & Hall/CRC, London, 1994).
Acknowledgements
This work was supported by Biotechnology and Biological Sciences Research Council (UK) grant BB/N000609/1. F.F.N. was supported by a Royal Society and British Academy Newton International Fellowship (UK) grant number NF140338.
Author information
Authors and Affiliations
Contributions
F.F.N. conceived the idea. F.F.N., M.d.R. and Z.Y. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nascimento, F.F., Reis, M.d. & Yang, Z. A biologist’s guide to Bayesian phylogenetic analysis. Nat Ecol Evol 1, 1446–1454 (2017). https://doi.org/10.1038/s41559-017-0280-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0280-x
This article is cited by
-
Rediscovering the unusual, solitary bryozoan Monobryozoon ambulans Remane, 1936: first molecular and new morphological data clarify its phylogenetic position
Frontiers in Zoology (2024)
-
Isolation of urease-producing bacteria from cocoa farms soils in Santander, Colombia, for cadmium remediation
3 Biotech (2023)
-
Extensive Interspecific Gene Flow Shaped Complex Evolutionary History and Underestimated Species Diversity in Rapidly Radiated Dolphins
Journal of Mammalian Evolution (2022)