Abstract
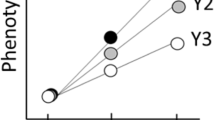
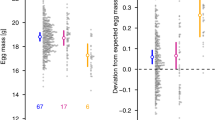
Male reproductive success depends on the competitive ability of sperm to fertilize the ova, which should lead to strong selection on sperm characteristics. This raises the question of how heritable variation in sperm traits is maintained. Here we show that in zebra finches (Taeniopygia guttata) nearly half of the variance in sperm morphology is explained by an inversion on the Z chromosome with a 40% allele frequency in the wild. The sperm of males that are heterozygous for the inversion had the longest midpieces and the highest velocity. Furthermore, such males achieved the highest fertility and the highest siring success, both within-pair and extra-pair. Males homozygous for the derived allele show detrimental sperm characteristics and the lowest siring success. Our results suggest heterozygote advantage as the mechanism that maintains the inversion polymorphism and hence variance in sperm design and in fitness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bennison, C., Hemmings, N., Slate, J. & Birkhead, T. Long sperm fertilize more eggs in a bird. Proc. R. Soc. B Biol. Sci. 282, 20141897 (2015).
Birkhead, T. R. & Pizzari, T. Postcopulatory sexual selection. Nat. Rev. Genet. 3, 262–273 (2002).
Parker, G. A. in Sperm Competition and Sexual Selection (eds Birkhead, T. R. & Møller, A. P.) 3–54 (Academic, 1998).
Pizzari, T. & Parker, G. A. in Sperm Biology: An Evolutionary Perspective (eds Birkhead, T. R. et al.) 207–245 (Elsevier, 2009).
Kleven, O., Laskemoen, T., Fossoy, F., Robertson, R. J. & Lifjeld, J. T. Intraspecific variation in sperm length is negatively related to sperm competition in passerine birds. Evolution 62, 494–499 (2008).
Lynch, M. & Walsh, B. Genetics and Analysis of Quantitative Traits (Sinauer, 1998).
Birkhead, T. R., Pellatt, E. J., Brekke, P., Yeates, R. & Castillo-Juarez, H. Genetic effects on sperm design in the zebra finch. Nature 434, 383–387 (2005).
Simmons, L. W. & Moore, A. J. in Sperm Biology: An Evolutionary Perspective (eds Birkhead, T. R. et al.) 401–430 (Elsevier, 2009).
Fisher, H. S., Jacobs-Palmer, E., Lassance, J.-M. & Hoekstra, H. E. The genetic basis and fitness consequences of sperm midpiece size in deer mice. Nat. Commun. 7, 13652 (2016).
Jamieson, B. G. M. in Reproductive Biology and Phylogeny of Birds Vol. 6A (ed. Jamieson, B. G. M.) 349–511 (CRC, 2007).
Immler, S. & Birkhead, T. R. Sperm competition and sperm midpiece size: no consistent pattern in passerine birds. Proc. R. Soc. B Biol. Sci. 274, 561–568 (2007).
Birkhead, T. R., Fletcher, F., Pellatt, E. J. & Staples, A. Ejaculate quality and the success of extra-pair copulations in the zebra finch. Nature 377, 422–423 (1995).
Bennison, C., Hemmings, N., Brookes, L., Slate, J. & Birkhead, T. Sperm morphology, adenosine triphosphate (ATP) concentration and swimming velocity: unexpected relationships in a passerine bird. Proc. R. Soc. B Biol. Sci. 283, 20161558 (2016).
Immler, S., Griffth, S. C., Zann, R. & Birkhead, T. R. Intra-specific variance in sperm morphometry: a comparison between wild and domesticated zebra finches Taeniopygia guttata. Ibis 154, 480–487 (2012).
Küpper, C. et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83 (2016).
Tuttle, E. M. et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26, 344–350 (2016).
White, M. J. D. Animal Cytology and Evolution (Cambridge Univ. Press, 1977).
Anton, E., Blanco, J., Egozcue, J. & Vidal, F. Sperm studies in heterozygote inversion carriers: a review. Cytogenet. Genome Res. 111, 297–304 (2005).
Morgan, D. T. A cytogenetic study of inversions in Zea mays. Genetics 35, 153–174 (1950).
Navarro, A. & Ruiz, A. On the fertility effects of pericentric inversions. Genetics 147, 931–933 (1997).
Roberts, P. A. A positive correlation between crossing over within heterozygous pericentric inversions and reduced egg hatch of Drosophila females. Genetics 56, 179–187 (1967).
Del Priore, L. & Pigozzi, M. I. Heterologous synapsis and crossover suppression in heterozygotes for a pericentric inversion in the zebra finch. Cytogenet. Genome Res. 147, 154–160 (2015).
Krimbas, C. B. & Powell, J. R. Drosophila Inversion Polymorphism (CRC, 1992).
Knief, U. et al. Fitness consequences of polymorphic inversions in the zebra finch genome. Genome Biol. 17, 199 (2016).
Serre, D., Nadon, R. & Hudson, T. J. Large-scale recombination rate patterns are conserved among human populations. Genome Res. 15, 1547–1552 (2005).
al Basatena, N. K. S., Hoggart, C. J., Coin, L. J. & O’Reilly, P. F. The effect of genomic inversions on estimation of population genetic parameters from SNP data. Genetics 193, 243–253 (2013).
Thompson, M. J. & Jiggins, C. D. Supergenes and their role in evolution. Heredity 113, 1–8 (2014).
Sturtevant, A. H. & Mather, E. The interrelations of inversions, heterosis and recombination. Am. Nat. 72, 447–452 (1938).
Kirkpatrick, M. How and why chromosome inversions evolve. PLoS Biol. 8, e1000501 (2010).
Joron, M. et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203–207 (2011).
Gromko, M. H. & Richmond, R. C. Modes of selection maintaining an inversion polymorphism in Drosophila paulistorum. Genetics 88, 357–366 (1978).
Kirkpatrick, M. & Barton, N. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434 (2006).
Lowry, D. B. & Willis, J. H. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500 (2010).
Christidis, L. Chromosomal evolution within the family Estrildidae (Aves) I. The Poephilae. Genetica 71, 81–97 (1986).
Itoh, Y., Kampf, K., Balakrishnan, C. N. & Arnold, A. P. Karyotypic polymorphism of the zebra finch Z chromosome. Chromosoma 120, 255–264 (2011).
Mossman, J., Slate, J., Humphries, S. & Birkhead, T. Sperm morphology and velocity are genetically codetermined in the zebra finch. Evolution 63, 2730–2737 (2009).
Birkhead, T. R., Martinez, J. G., Burke, T. & Froman, D. P. Sperm mobility determines the outcome of sperm competition in the domestic fowl. Proc. R. Soc. B Biol. Sci. 266, 1759–1764 (1999).
Denk, A. G., Holzmann, A., Peters, A., Vermeirssen, E. L. M. & Kempenaers, B. Paternity in mallards: effects of sperm quality and female sperm selection for inbreeding avoidance. Behav. Ecol. 16, 825–833 (2005).
Ellegren, H. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 21, 2082–2086 (2011).
Arunkumar, K. P., Mita, K. & Nagaraju, J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182, 493–501 (2009).
Kim, K.-W. et al. A sex-linked supergene controls sperm morphology and swimming speed in a songbird. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-017-0235-2 (2017).
Forstmeier, W. Do individual females differ intrinsically in their propensity to engage in extra-pair copulations? PLoS ONE 2, e952 (2007).
Ihle, M., Kempenaers, B. & Forstmeier, W. Fitness benefits of mate choice for compatibility in a socially monogamous species. PLoS Biol. 13, e1002248 (2015).
Silcox, A. P. & Evans, S. M. Factors affecting the formation and maintenance of pair bonds in the zebra finch, Taeniopygia guttata. Anim. Behav. 30, 1237–1243 (1982).
Birkhead, T. R., Burke, T., Zann, R., Hunter, F. M. & Krupa, A. P. Extra-pair paternity and intraspecific brood parasitism in wild zebra finches Taeniopygia guttata, revealed by DNA fingerprinting. Behav. Ecol. Sociobiol. 27, 315–324 (1990).
Griffith, S. C., Holleley, C. E., Mariette, M. M., Pryke, S. R. & Svedin, N. Low level of extrapair parentage in wild zebra finches. Anim. Behav. 79, 261–264 (2010).
Tschirren, B., Postma, E., Rutstein, A. N. & Griffith, S. C. When mothers make sons sexy: maternal effects contribute to the increased sexual attractiveness of extra-pair offspring. Proc. R. Soc. B Biol. Sci. 279, 1233–1240 (2012).
Hooper, D. M. & Price, T. D. Rates of karyotypic evolution in Estrildid finches differ between island and continental clades. Evolution 69, 890–903 (2015).
Christidis, L. Chromosomal evolution within the family Estrildidae (Aves) II. The Lonchurae. Genetica 71, 99–113 (1986).
Chen, Z. J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14, 471–482 (2013).
Burley, N. T., Enstrom, D. A. & Chitwood, L. Extra-pair relations in zebra finches: differential male success results from female tactics. Anim. Behav. 48, 1031–1041 (1994).
Burley, N. T., Parker, P. G. & Lundy, K. Sexual selection and extrapair fertilization in a socially monogamous passerine, the zebra finch (Taeniopygia guttata). Behav. Ecol. 7, 218–226 (1996).
Forstmeier, W., Martin, K., Bolund, E., Schielzeth, H. & Kempenaers, B. Female extrapair mating behavior can evolve via indirect selection on males. Proc. Natl Acad. Sci. USA 108, 10608–10613 (2011).
Forstmeier, W., Segelbacher, G., Mueller, J. C. & Kempenaers, B. Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 16, 4039–4050 (2007).
Mathot, K. J., Martin, K., Kempenaers, B. & Forstmeier, W. Basal metabolic rate can evolve independently of morphological and behavioural traits. Heredity 111, 175–181 (2013).
Opatová, P. et al. Inbreeding depression of sperm traits in the zebra finch Taeniopygia guttata. Ecol. Evol. 6, 295–304 (2016).
Knief, U. et al. Quantifying realized inbreeding in wild and captive animal populations. Heredity 114, 397–403 (2015).
Gabriel, S., Ziaugra, L. & Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum Genet. Ch. 2, Unit 2.12 (2009).
Knief, U. et al. Association mapping of morphological traits in wild and captive zebra finches: reliable within but not between populations. Mol. Ecol. 26, 1285–1305 (2017).
Warren, W. C. et al. The genome of a songbird. Nature 464, 757–762 (2010).
Opatová, P. et al. Data from: Inbreeding depression of sperm traits in the zebra finch Taeniopygia guttata. Dryad Data Repository https://doi.org/10.5061/dryad.4h245 (2016).
Cramer, E. R. A., Ålund, M., McFarlane, S. E., Johnsen, A. & Qvarnström, A. Females discriminate against heterospecific sperm in a natural hybrid zone. Evolution 70, 1844–1855 (2016).
Laskemoen, T. et al. Sperm quantity and quality effects on fertilization success in a highly promiscuous passerine, the tree swallow Tachycineta bicolor. Behav. Ecol. Sociobiol. 64, 1473–1483 (2010).
Knief, U., Schielzeth, H., Ellegren, H., Kempenaers, B. & Forstmeier, W. A prezygotic transmission distorter acting equally in female and male zebra finches Taeniopygia guttata. Mol. Ecol. 24, 3846–3859 (2015).
Schielzeth, H. & Bolund, E. Patterns of conspecific brood parasitism in zebra finches. Anim. Behav. 79, 1329–1337 (2010).
Forstmeier, W., Schielzeth, H., Schneider, M. & Kempenaers, B. Development of polymorphic microsatellite markers for the zebra finch (Taeniopygia guttata). Mol. Ecol. Notes 7, 1026–1028 (2007).
Wang, D., Kempenaers, N., Kempenaers, B. & Forstmeier, W. Male zebra finches have limited ability to identify high-fecundity females. Behav. Ecol. 28, 784–792 (2017).
Backström, N. et al. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20, 485–495 (2010).
Forstmeier, W., Wagenmakers, E. J. & Parker, T. H. Detecting and avoiding likely false-positive findings—a practical guide. Biol. Rev. https://doi.org/10.1111/brv.12315 (2016).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Vazquez, A. I., Bates, D. M., Rosa, G. J. M., Gianola, D. & Weigel, K. A. Technical note: an R package for fitting generalized linear mixed models in animal breeding. J. Anim. Sci. 88, 497–504 (2010).
Nakagawa, S. & Schielzeth, H. A general and simple method for obtaining R 2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142 (2013).
Bartoń, K. MuMIn: Multi-model inference. R package v. 1.15.6 (2016).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S 4th edn (Springer, 2002).
Gilmour, A. R., Gogel, B. J., Cullis, B. R. & Thompson, R. Asreml User Guide Release 3.0 (VSN International, 2009).
Wolak, M. E. nadiv: an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol. Evol. 3, 792–796 (2012).
Kruuk, L. E. B. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. Lond. B Biol. Sci. 359, 873–890 (2004).
Kruuk, L. E. B. & Hadfield, J. D. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 20, 1890–1903 (2007).
Zuur, A. F., Ieno, E. N. & Elphick, C. S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 1, 3–14 (2010).
Kinghorn, B. P. & Kinghorn, A. J. Pedigree Viewer 6.5 (Univ. New England, 2010).
Acknowledgements
We thank T. Aronson, E. Bolund, S. Janker, H. Schielzeth, J. Schreiber and O. Tomášek for help with data collection, M. Schneider and G. Hemmrich-Stanisak for molecular and genomic work, and S. Bauer, E. Bodendorfer, J. Didsbury, A. Grötsch, A. Kortner, P. Neubauer, F. Weigel and B. Wörle for animal care and help with breeding. This work was supported by the Max Planck Society (B.K.) and by the Czech Science Foundation (project no. P506/12/2472 to T.A.).
Author information
Authors and Affiliations
Contributions
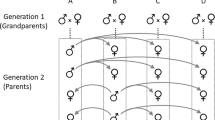
U.K., M.W. and A.F. genotyped all birds. T.A., J.A. and K.M. collected sperm samples. P.O. measured sperm morphology. J.A. measured sperm velocity. W.F., M.I., D.W. and K.M. collected breeding data. U.K., W.F. and Y.P. analysed the data. U.K., W.F. and B.K. wrote the manuscript with help from T.A. All authors contributed to the final manuscript. W.F., T.A. and B.K. conceived of the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures 1–4, Supplementary Tables 1–10, Supplementary References
Rights and permissions
About this article
Cite this article
Knief, U., Forstmeier, W., Pei, Y. et al. A sex-chromosome inversion causes strong overdominance for sperm traits that affect siring success. Nat Ecol Evol 1, 1177–1184 (2017). https://doi.org/10.1038/s41559-017-0236-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0236-1
This article is cited by
-
Intralocus conflicts associated with a supergene
Nature Communications (2022)
-
Impact of male trait exaggeration on sex-biased gene expression and genome architecture in a water strider
BMC Biology (2021)
-
Mutation load at a mimicry supergene sheds new light on the evolution of inversion polymorphisms
Nature Genetics (2021)
-
Reproductive coordination breeds success: the importance of the partnership in avian sperm biology
Behavioral Ecology and Sociobiology (2020)
-
Postcopulatory sexual selection reduces Z-linked genetic variation and might contribute to the large Z effect in passerine birds
Heredity (2019)