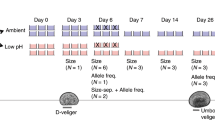
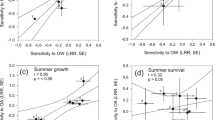
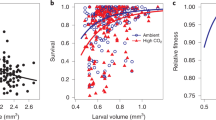
Abstract
Predicting the response of marine animals to climate change is hampered by a lack of multigenerational studies on evolutionary adaptation, particularly to combined ocean warming and acidification (OWA). We provide evidence for rapid adaptation to OWA in the foundational copepod species, Acartia tonsa, by assessing changes in population fitness on the basis of a comprehensive suite of life-history traits, using an orthogonal experimental design of nominal temperature (18 °C, 22 °C) and \(p_{\mathrm{{CO}}_2}\) (400, 2,000 µatm) for 25 generations (~1 year). Egg production and hatching success initially decreased under OWA, resulting in a 56% reduction in fitness. However, both traits recovered by the third generation, and average fitness was reduced thereafter by only 9%. Antagonistic interactions between warming and acidification in later generations decreased survival, thereby limiting full fitness recovery. Our results suggest that such interactions constrain evolutionary rescue and add complexity to predictions of the responses of animal populations to climate change.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The phenotypic and physical data referred to in the text are deposited in Zenodo (https://doi.org/10.5281/zenodo.5119920)75. The genetic diversity data are deposited in GenBank: BioProject number PRJNA590963. Source data are provided with this paper.
Code availability
The scripts for analysis of the physical, phenotypic and genetic diversity data are deposited in Zenodo (https://doi.org/10.5281/zenodo.5119920).
References
Hönisch, B. et al. The geological record of ocean acidification. Science 335, 1058–1063 (2012).
Bindoff, N. L. et al. in Special Report on the Ocean and Cryosphere in a Changing Climate (eds Pörtner, H.-O. et al.) 447–588 (IPCC, 2019).
Pörtner, H.-O. et al. in Special Report on the Ocean and Cryosphere in a Changing Climate (eds Pörtner, H.-O. et al.) 35–74 (IPCC, 2019).
Caldeira, K. & Wickett, M. E. Anthropogenic carbon and ocean pH. Nature 425, 365 (2003).
Cai, W. J. et al. Acidification of subsurface coastal waters enhanced by eutrophication. Nat. Geosci. 4, 766–770 (2011).
Wallace, R. B., Baumann, H., Grear, J. S., Aller, R. C. & Gobler, C. J. Coastal ocean acidification: the other eutrophication problem. Estuar. Coast. Shelf Sci. 148, 1–13 (2014).
Munday, P. L., Warner, R. R., Monro, K., Pandolfi, J. M. & Marshall, D. J. Predicting evolutionary responses to climate change in the sea. Ecol. Lett. 16, 1488–1500 (2013).
Schlichting, C. D. & Pigliucci, M. Phenotypic Evolution: A Reaction Norm Perspective (Sinauer Associates, 1998).
Kelly, M. W. & Hofmann, G. E. Adaptation and the physiology of ocean acidification. Funct. Ecol. 27, 980–990 (2013).
Pespeni, M. H. et al. Evolutionary change during experimental ocean acidification. Proc. Natl Acad. Sci. USA 110, 6937–6942 (2013).
Thor, P. & Dupont, S. Transgenerational effects alleviate severe fecundity loss during ocean acidification in a ubiquitous planktonic copepod. Glob. Change Biol. 21, 2261–2271 (2015).
Donelson, J. M., Salinas, S., Munday, P. L. & Shama, L. N. S. Transgenerational plasticity and climate change experiments: where do we go from here? Glob. Change Biol. 24, 13–34 (2018).
Chevin, L. M., Lande, R. & Mace, G. M. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8, e1000357 (2010).
Angilletta, M. J. Thermal Adaptation: A Theoretical and Empirical Synthesis (Oxford University Press, 2009).
Byrne, M. in Oceanography and Marine Biology: An Annual Review Vol. 49 (eds Gibson, R. N. et al.) Ch. 1 (CRC Press, 2011).
Whiteley, N. M. Physiological and ecological responses of crustaceans to ocean acidification. Mar. Ecol. Prog. Ser. 430, 257–271 (2011).
Cripps, G., Lindeque, P. & Flynn, K. J. Have we been underestimating the effects of ocean acidification in zooplankton? Glob. Change Biol. 20, 3377–3385 (2014).
Baumann, H. Experimental assessments of marine species sensitivities to ocean acidification and co-stressors: how far have we come? Can. J. Zool. 97, 399–408 (2019).
Gibbin, E. M. et al. Can multi-generational exposure to ocean warming and acidification lead to the adaptation of life history and physiology in a marine metazoan? J. Exp. Biol. 220, 551–563 (2017).
Gibbin, E. M., Massamba N’Siala, G., Chakravarti, L. J., Jarrold, M. D. & Calosi, P. The evolution of phenotypic plasticity under global change. Sci. Rep. 7, 17253 (2017).
Gonzalez, A., Ophelie, R., Ferriere, R. & Hochberg, M. E. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Philos. Trans. R. Soc. Lond. B 368, 20120404 (2012).
Bell, G. & Gonzalez, A. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948 (2009).
Carlson, S. M., Cunningham, C. J. & Westley, P. A. H. Evolutionary rescue in a changing world. Trends Ecol. Evol. 29, 521–530 (2014).
Hardy, A. The Open Sea: The World of Plankton (Fontana Collins, 1970).
Huys, R. & Boxshall, G. A. Copepod Evolution (The Ray Society, 1991).
Beaugrand, G. & Reid, P. C. Long-term changes in phytoplankton, zooplankton and salmon related to climate. Glob. Change Biol. 9, 801–817 (2003).
Möllmann, C., Müller-Karulis, B., Kornilovs, G. & St John, M. A. Effects of climate and overfishing on zooplankton dynamics and ecosystem structure: regime shifts, trophic cascade, and feedback loops in a simple ecosystem. ICES J. Mar. Sci. 65, 302–310 (2008).
Steinberg, D. K. & Landry, M. R. Zooplankton and the ocean carbon cycle. Annu. Rev. Mar. Sci. 9, 413–444 (2017).
Mauchline, J. (ed.) The Biology of Calanoid Copepods (Academic Press, 1998).
Turner, J. T. The Feeding Ecology of Some Zooplankters That Are Important Prey Items of Larval Fish. NOAA NMFS Technical Report (1984).
Rice, E., Dam, H. G. & Stewart, G. Impact of climate change on estuarine zooplankton: surface water warming in Long Island Sound is associated with changes in copepod size and community structure. Estuaries Coast 38, 13–23 (2015).
Gobler, C. J. & Baumann, H. Hypoxia and acidification in marine ecosystems: coupled dynamics and effects on ocean life. Biol. Lett. 12, 20150976 (2016).
Côté, I. M., Darling, E. S. & Brown, C. J. Interactions among ecosystem stressors and their importance in conservation. Proc. R. Soc. Lond. B 283, 20152592 (2016).
Burt, A. Perspective: the evolution of fitness. Evolution 49, 1–8 (1995).
Hendry, A. P. & Gonzalez, A. Whither adaptation? Biol. Philos. 23, 673–699 (2008).
Arnold, S. J., Pfrender, M. E. & Jones, A. G. The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica 112–113, 9–32 (2001).
Caswell, H. Matrix Population Models: Construction, Analysis, and Interpretation (Sinauer Associates, 2001).
Sasaki, M. C. & Dam, H. G. Integrating patterns of thermal tolerance and phenotypic plasticity with population genetics to improve understanding of vulnerability to warming in a widespread copepod. Glob. Change Biol. 25, 4147–4164 (2019).
Luikart, G., England, P. R., Tallmon, D., Jordan, S. & Taberlet, P. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4, 981–994 (2003).
Black, W. C. IV, Baer, C. F., Antolin, M. F. & DuTeau, N. M. Population genomics: genome-wide sampling of insect populations. Annu. Rev. Entomol. 46, 441–469 (2001).
Brennan, R. et al. Loss and recovery of transcriptional plasticity after long-term adaptation to global change conditions in a marine copepod. Preprint at bioRxiv https://doi.org/10.1101/2020.01.29.925396 (2020).
Kingsolver, J. G. & Pfennig, D. W. Patterns and power of phenotypic selection in nature. Bioscience 57, 561–572 (2007).
Crespi, B. J. & Bookstein, F. L. A path-analytic model for the measurement of selection on morphology. Evolution 43, 18–28 (1989).
Pigliucci, M. & Kaplan, J. Making Sense of Evolution (Univ. Chicago Press, 2006); https://doi.org/10.7208/chicago/9780226668352.001.0001
Bush, A. et al. Incorporating evolutionary adaptation in species distribution modelling reduces projected vulnerability to climate change. Ecol. Lett. 19, 1468–1478 (2016).
Riebesell, U. & Gattuso, J. Lessons learned from ocean acidification research. Nat. Clim. Change 5, 2014–2016 (2015).
Langer, J. A. F., Meunier, C. L., Ecker, U. & Horn, H. G. Acclimation and adaptation of the coastal calanoid copepod Acartia tonsa to ocean acidification: a long-term laboratory investigation. Mar. Ecol. Prog. Ser. 619, 35–51 (2019).
De Wit, P., Dupont, S. & Thor, P. Selection on oxidative phosphorylation and ribosomal structure as a multigenerational response to ocean acidification in the common copepod Pseudocalanus acuspes. Evol. Appl. 9, 1112–1123 (2016).
Chakravarti, L. J. et al. Can trans-generational experiments be used to enhance species resilience to ocean warming and acidification? Evol. Appl. 9, 1133–1146 (2016).
Carrier-Belleau, C., Drolet, D., McKindsey, C. W. & Archambault, P. Environmental stressors, complex interactions and marine benthic communities’ responses. Sci. Rep. 11, 4194 (2021).
Dam, H. G. & Baumann, H. in Climate Change Impacts on Fisheries and Aquaculture: A Global Analysis (eds Phillips, B. F. and Pérez-Ramírez, M.) 851–874 (Wiley, 2017).
Bell, G. Evolutionary rescue and the limits of adaptation. Philos. Trans. R. Soc. Lond. B 368, 20120080 (2013).
Falconer, D. S. Introduction to Quantitative Genetics (Longman Scientific and Technical, 1989).
Angilletta, M. J. Jr Estimating and comparing thermal performance curves. J. Therm. Biol. 31, 541–545 (2006).
Feinberg, L. R. & Dam, H. G. Effects of diet on dimensions, density and sinking rates of fecal pellets of the copepod Acartia tonsa. Mar. Ecol. Prog. Ser. 175, 87–96 (1998).
Pierrot, D., Lewis, E. & Wallace, D. W. R. MS Excel Program Developed for CO2 System Calculations. ORNL/CDIAC-105a. (Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, 2006); https://doi.org/10.3334/CDIAC/otg.CO2SYS_XLS_CDIAC105a
Lueker, T. J., Dickson, A. G. & Keeling, C. D. Ocean \(p_{{\mathrm{CO}}_2}\) calculated from dissolved inorganic carbon, alkalinity, and equations for K1 and K2: validation based on laboratory measurements of CO2 in gas and seawater at equilibrium. Mar. Chem. 70, 105–119 (2000).
Dickson, A. G. Standard potential of the reaction: AgCl(s) + 12H2 (g) = Ag(s) + HCl (aq), and the standard acidity constant of the ion HSO4– in synthetic sea water from 273.15 to 318.15 K. J. Chem. Thermodyn. 22, 113–127 (1990).
Uppström, L. R. The boron/chlorinity ratio of deep-sea water from the Pacific Ocean. Deep Sea Res. Oceanogr. Abstr. 21, 161–162 (1974).
Murray, C. S. & Baumann, H. You better repeat it: complex CO2× temperature effects in Atlantic silverside offspring revealed by serial experimentation. Diversity 10, 69 (2018).
Schank, J. C. & Koehnle, T. J. Pseudoreplication is a Pseudoproblem. J. Comp. Psychol. 123, 421–433 (2009).
Oksanen, L. Logic of experiments in ecology: is pseudoreplication a pseudoissue? Oikos 94, 27–38 (2001).
Therneau, T. A Package for Survival Analysis in R. R package 3.2-11 (2021); https://CRAN.R-project.org/package=survival
Lande, R. & Arnold, S. J. The measurement of selection on correlated characters. Evolution 37, 1210–1226 (1983).
Rosseel, Y. lvaan: an R package for structural equation modeling. J. Stat. Softw. https://doi.org/10.18637/jss.v048.i02 (2012).
Epskamp, S., Stuber, S., Nak, J., Veenman, M. & Jorgensen, T. D. semPlot: Path Diagrams and Visual Analysis of Various SEM Packages’ Output. (2019); https://CRAN.R-project.org/package=semPlot
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Jørgensen, T. S. et al. The genome and mRNA transcriptome of the cosmopolitan calanoid copepod Acartia tonsa Dana improve the understanding of copepod genome size evolution. Genome Biol. Evol. 11, 1440–1450 (2019).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kofler, R. et al. Popoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6, e15925 (2011).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2020); https://www.R-project.org/
Wood, S. N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 73, 3–36 (2011).
Simpson, G. L. Modelling palaeoecological time series using generalised additive models. Front. Ecol. Evol. 6, 149 (2018).
Dam, H. G. et al. Data and code repository for ‘Rapid, but limited, zooplankton adaptation to simultaneous warming and acidification’. Zenodo https://doi.org/10.5281/zenodo.5115103 (2021).
Acknowledgements
Research was supported by grants from the USA National Science Foundation (OCE-1559180 awarded to H.G.D., M.B.F. and H.B.; and OCE-1559075 awarded to M.H.P.) and Connecticut Sea Grant (R/LR‐25) awarded to H.G.D., M.B.F. and H.B. The authors thank W. Huffman for aiding in pilot experiments; C. Murray for assistance in alkalinity measurements; D. Arbige, C. Woods and B. Dziomba for help in maintaining equipment and constructing custom enclosures for the experiments; and T. Moore and J. Lee of UConn’s Statistical Consulting Services for advice and assistance on data analysis.
Author information
Authors and Affiliations
Contributions
H.G.D. conceived the project, designed research, aided in data analysis and wrote the manuscript. J.A.deM. conducted experiments, analysed data, created figures and wrote the manuscript with H.G.D. G.P., L.N. and X.H. conducted experiments. M.B.F. conceived the project and designed research. H.B. conceived the project, designed research and designed the CO2 delivery system. R.S.B. performed genomic diversity analysis. M.H.P. conceived the project, designed research and performed genomic analysis. All authors edited and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Climate Change thanks Peter Thor and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Development time vs generation for transgenerational study.
Shown are the mean calculated development times (naupliar stage 1 to adult) for each treatment at each generation where life-history traits are measured. Curves for treatments are offset for clarity. Treatment colors: blue: AM; green: OA; orange: OW; brown: OWA. Box and whisker plots for these data are available in Supplemental Fig. 4.
Extended Data Fig. 2 Sex ratio vs generation for transgenerational study.
Results for sex ratio across generations modeled as A) linear model and B) Generalized Additive Model. Treatment colors: blue: AM; green: OA; orange: OW; brown: OWA.
Extended Data Fig. 3 Frequency distribution of population fitness values (λ) for the four treatments in the transgenerational experiment.
Treatment colors: blue: AM; green: OA; orange: OW; brown: OWA.
Extended Data Fig. 4 Predicted probabilities of non-zero fitness (lambda) values vs generations across treatments in the transgenerational experiment.
Shown are predicted mean non-zero lambda probabilities. Probabilities for ambient (AM), ocean acidification (OA), and ocean warming (OW) treatments are statistically independent of generations. Probabilities for the simultaneous ocean warming and acidification (OWA) significantly increase with generation. Shading represents 95% confidence intervals around the mean. Treatment colors: blue: AM; green: OA; orange: OW; brown: OWA.
Extended Data Fig. 5 Estimates of genetic diversity (π) at generation 25 vs treatments of the transgenerational experiment.
Estimates were calculated in 100 bp non-overlapping sliding windows. Windows were included when at least 50% of sites had coverage between 30x and 1000x per sample and the window was covered across all samples. The asterisk indicates the sample in the OA treatment with reduced genetic diversity relative to other samples (Wilcoxon Rank Sum test with Holm correction for multiple testing; p < 0.05); all other samples were not significantly different (p > 0.05). In the boxes, the centre black line represents the median, the circles represent means, upper box edge represents the 75% quartile, lower box edge represents 25% quartile, whiskers represent 1.5x interquartile range, and points represent outliers. Treatment colors: blue: AM; green: OA; orange: OW; brown: OWA.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Tables 1–5.
Source data
Source Data Fig. 1
Unprocessed values of egg production and hatching success.
Source Data Fig. 2
Unprocessed values for survival.
Source Data Fig. 3
Unprocessed values for fitness (λ).
Source Data Fig. 4
Sheet 1: Table of fitness (λ) and relative fitness values with corresponding survival, egg production, hatching success, development time and sex ratio values for the first and last evaluated generations. Sheet 2: Table of survival values for the first and last generations.
Source Data Extended Data Fig. 1
Unprocessed values for calculated development time.
Source Data Extended Data Fig. 2
Unprocessed values for observed sex ratio (proportion of females relative to males).
Source Data Extended Data Fig. 3
Unprocessed values for fitness (λ).
Source Data Extended Data Fig. 4
Unprocessed values for fitness (λ) with binary transformed λ. λ values > 0 are given a binary value of 1, and λ values = 0 are given a binary value of 0.
Source Data Extended Data Fig. 5
Unprocessed values of nucleotide diversity for F25.
Rights and permissions
About this article
Cite this article
Dam, H.G., deMayo, J.A., Park, G. et al. Rapid, but limited, zooplankton adaptation to simultaneous warming and acidification. Nat. Clim. Chang. 11, 780–786 (2021). https://doi.org/10.1038/s41558-021-01131-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41558-021-01131-5
This article is cited by
-
Steeper size spectra with decreasing phytoplankton biomass indicate strong trophic amplification and future fish declines
Nature Communications (2024)
-
Monitoring and modelling marine zooplankton in a changing climate
Nature Communications (2023)
-
Epigenetic plasticity enables copepods to cope with ocean acidification
Nature Climate Change (2022)
-
Upper environmental pCO2 drives sensitivity to ocean acidification in marine invertebrates
Nature Climate Change (2022)
-
Loss of transcriptional plasticity but sustained adaptive capacity after adaptation to global change conditions in a marine copepod
Nature Communications (2022)