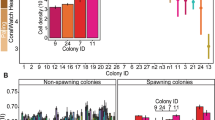
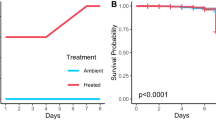
Abstract
The perception that the inheritance of phenotypic traits operates solely through genetic means is slowly being eroded: epigenetic mechanisms have been shown to induce heritable changes in gene activity in plants1,2 and metazoans1,3. Inheritance of DNA methylation patterns provides a potential pathway for environmentally induced phenotypes to contribute to evolution of species and populations1,2,3,4,5. However, in basal metazoans, it is unknown whether inheritance of CpG methylation patterns occurs across the genome (as in plants) or as rare exceptions (as in mammals)4. Here, we show that DNA methylation patterns in a reef-building coral are determined by genotype and developmental stage, as well as by parental environment. Transmission of CpG methylation from adults to their sperm and larvae demonstrates genome-wide inheritance. Variation in the hypermethylation of genes in adults and their sperm from distinct environments suggests intergenerational acclimatization to local temperature and salinity. Furthermore, genotype-independent adjustments of methylation levels in stress-related genes were strongly correlated with offspring survival rates under heat stress. These findings support a role of DNA methylation in the intergenerational inheritance of traits in corals, which could extend to enhancing their capacity to adapt to climate change.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Whole-genome bisulfite sequencing data can be found in NCBI BioProject PRJNA430328. Individual SRA accessions are listed in Supplementary Dataset 1c. Genomic sequences and annotations are available at http://pdae.reefgenomics.org/ (ref. 55).
Code availability
Scripts used to analyse methylation data (and the underlying theoretical justifications) are detailed at https://github.com/lyijin/working_with_dna_meth. Other analytical and plotting scripts, key intermediate files and further explanatory notes are available at https://doi.org/10.5281/zenodo.3558476 (v.1.0.0).
References
Jablonka, E. & Raz, G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176 (2009).
Lamke, J. & Baurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017).
Lim, J. P. & Brunet, A. Bridging the transgenerational gap with epigenetic memory. Trends Genet. 29, 176–186 (2013).
Heard, E. & Martienssen, R. A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 (2014).
Eirin-Lopez, J. M. & Putnam, H. M. Marine environmental epigenetics. Annu. Rev. Mar. Sci. 11, 335–368 (2019).
Moberg, F. & Folke, C. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233 (1999).
Hughes, T. P. et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83 (2018).
Hughes, T. P. et al. Global warming and recurrent mass bleaching of corals. Nature 543, 373–377 (2017).
van Oppen, M. J., Oliver, J. K., Putnam, H. M. & Gates, R. D. Building coral reef resilience through assisted evolution. Proc. Natl Acad. Sci. USA 112, 2307–2313 (2015).
Torda, G. et al. Rapid adaptive responses to climate change in corals. Nat. Clim. Change 7, 627–636 (2017).
Liew, Y. J. et al. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci. Adv. 4, eaar8028 (2018).
Dixon, G., Liao, Y., Bay, L. K. & Matz, M. V. Role of gene body methylation in acclimatization and adaptation in a basal metazoan. Proc. Natl Acad. Sci. USA 115, 13342–13346 (2018).
Li, Y. et al. DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci. Adv. 4, eaat2142 (2018).
Howells, E. J. et al. Host adaptation and unexpected symbiont partners enable reef-building corals to tolerate extreme temperatures. Glob. Change Biol. 22, 2702–2714 (2016).
Howells, E. J. et al. Species-specific trends in the reproductive output of corals across environmental gradients and bleaching histories. Mar. Pollut. Bull. 105, 532–539 (2016).
Riegl, B. et al. Population collapse dynamics in Acropora downingi, an Arabian/Persian Gulf ecosystem-engineering coral, linked to rising temperature. Glob. Change Biol. 24, 2447–2462 (2018).
Dixon, G. B., Bay, L. K. & Matz, M. V. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genom. 15, 1109 (2014).
Dimond, J. L. & Roberts, S. B. Germline DNA methylation in reef corals: patterns and potential roles in response to environmental change. Mol. Ecol. 25, 1895–1904 (2016).
Wang, X. et al. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9, e1003872 (2013).
Lyko, F. et al. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 (2010).
Glastad, K. M., Gokhale, K., Liebig, J. & Goodisman, M. A. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 6, 37110 (2016).
Rosic, N. N. & Dove, S. Mycosporine-like amino acids from coral dinoflagellates. Appl. Environ. Microbiol. 77, 8478–8486 (2011).
Imbs, A. B. & Yakovleva, I. M. Dynamics of lipid and fatty acid composition of shallow-water corals under thermal stress: an experimental approach. Coral Reefs 31, 41–53 (2012).
Tolosa, I., Treignier, C., Grover, R. & Ferrier-Pages, C. Impact of feeding and short-term temperature stress on the content and isotopic signature of fatty acids, sterols, and alcohols in the scleractinian coral Turbinaria reniformis. Coral Reefs 30, 763–774 (2011).
Hillyer, K. E., Tumanov, S., Villas-Boas, S. & Davy, S. K. Metabolite profiling of symbiont and host during thermal stress and bleaching in a model cnidarian–dinoflagellate symbiosis. J. Exp. Biol. 219, 516–527 (2016).
Ogawa, Y. & Imamoto, N. Nuclear transport adapts to varying heat stress in a multistep mechanism. J. Cell Biol. 217, 2341–2352 (2018).
Kosako, H. et al. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16, 1026–1035 (2009).
Downs, C. A. et al. Cellular pathology and histopathology of hypo-salinity exposure on the coral Stylophora pistillata. Sci. Total Environ. 407, 4838–4851 (2009).
Spector, A. A. Arachidonic acid cytochrome P450 epoxygenase pathway. J. Lipid Res. 50, S52–S56 (2009).
Vin, H. et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. eLife 2, e00969 (2013).
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K. L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008).
Grunewald, S., Paasch, U., Glander, H. J. & Anderegg, U. Mature human spermatozoa do not transcribe novel RNA. Andrologia 37, 69–71 (2005).
Zimmer, R. K. & Riffell, J. A. Sperm chemotaxis, fluid shear, and the evolution of sexual reproduction. Proc. Natl Acad. Sci. USA 108, 13200–13205 (2011).
Nakamura, N. Ubiquitination regulates the morphogenesis and function of sperm organelles. Cells 2, 732–750 (2013).
Song, W. H., Yi, Y. J., Sutovsky, M., Meyers, S. & Sutovsky, P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl Acad. Sci. USA 113, E5261–E5270 (2016).
Putnam, H. M., Davidson, J. M. & Gates, R. D. Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178 (2016).
Hughes, T. P. et al. Ecological memory modifies the cumulative impact of recurrent climate extremes. Nat. Clim. Change 9, 40–43 (2019).
Middlebrook, R., Hoegh-Guldberg, O. & Leggat, W. The effect of thermal history on the susceptibility of reef-building corals to thermal stress. J. Exp. Biol. 211, 1050–1056 (2008).
Brown, B. E., Dunne, R. P., Edwards, A. J., Sweet, M. J. & Phongsuwan, N. Decadal environmental ‘memory’ in a reef coral? Mar. Biol. 162, 479–483 (2015).
Bellantuono, A. J., Granados-Cifuentes, C., Miller, D. J., Hoegh-Guldberg, O. & Rodriguez-Lanetty, M. Coral thermal tolerance: tuning gene expression to resist thermal stress. PLoS ONE 7, e50685 (2012).
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N. & Bay, R. A. Mechanisms of reef coral resistance to future climate change. Science 344, 895–898 (2014).
Hofmeister, B. T., Lee, K., Rohr, N. A., Hall, D. W. & Schmitz, R. J. Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 18, 155 (2017).
Lauss, K. et al. Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 176, 1627–1645 (2018).
van Oppen, M. J. H. et al. Shifting paradigms in restoration of the world’s coral reefs. Glob. Change Biol. 23, 3437–3448 (2017).
Howells, E. J., Abrego, D., Vaughan, G. O. & Burt, J. A. Coral spawning in the Gulf of Oman and relationship to latitudinal variation in spawning season in the northwest Indian Ocean. Sci. Rep. 4, 7484 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Liu, Y., Siegmund, K. D., Laird, P. W. & Berman, B. P. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 13, R61 (2012).
Bhatia, G., Patterson, N., Sankararaman, S. & Price, A. L. Estimating and interpreting FST: the impact of rare variants. Genome Res. 23, 1514–1521 (2013).
Shapiro, S. S. & Wilk, M. B. An analysis of variance test for normality (Complete Samples). Biometrika 52, 591–611 (1965).
Levene, H. in Contributions to Probability and Statistics Vol. 1 (ed. Olkin, I.) 278–292 (Stanford Univ. Press, 1960).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Benjamini, Y. & Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001).
Alexa, A., Rahnenfuhrer, J. & Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006).
Liew, Y. J., Aranda, M. & Voolstra, C. R. Reefgenomics.Org—a repository for marine genomics data. Database 2016, baw152 (2016).
Acknowledgements
We thank D. Abrego, G. Vaughan and D. McParland for assistance with fieldwork, coral spawning and the collection of environmental data. We thank the NYUAD Core Research Vessel and The Palms Dive Center for fieldwork support. We thank the Environment Agency Abu Dhabi and Fujairah Municipality for research permits and the KAUST Sequencing Core Facility for the sequencing of the libraries. The research reported in this publication was supported by the KAUST OSR under grant no. URF/1/3447-01-01, as well as baseline support to M.A.; and by NYUAD research grant no. AD105 to Y.I.
Author information
Authors and Affiliations
Contributions
E.J.H, Y.I. and M.A. conceived and coordinated the project. M.A., X.W. and J.A.B provided resources. E.J.H. collected samples from the wild, performed controlled crosses and extracted DNA from fixed samples. C.T.M. constructed libraries for WGBS and RNA-seq. Y.J.L., Y.I. and M.A. analysed data. Y.J.L., E.J.H. and M.A. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Climate Change thanks Eva Majerová and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Methylation in P. daedalea is more commonly found in genic regions, and concentrated closer to the 5’ and 3’ ends.
(a) Genic regions are significantly more frequently methylated than intergenic regions (3.9% versus 3.0%; Fisher’s exact P < 10-300). (b) Methylation levels are bimodally distributed in exons, introns and intergenic regions. Exons have the highest methylation levels, followed by introns and intergenic regions. (c) Relative frequencies of methylated positions across a standardized gene model with flanking 4 kb regions indicate that methylated positions are more frequently found at both ends of the gene model. Solid lines depict transcriptional start site (left) and transcription termination site (right) while dotted lines delineate the borders of the indicated genomic feature. The widths of the features correspond to mean normalized lengths of the respective exons and introns in P. daedalea (exons, from left to right: 286 bp, 320 bp, 225 bp, 203 bp, 270 bp, 380 bp; introns, from left to right: 1,971 bp, 1,744 bp, 1,598 bp, 1,728 bp).
Extended Data Fig. 2 Per-origin principal components analysis of DNA methylation patterns in P. daedalea.
Plots show variation explained by the first three principal components of PCAs carried out separately on samples from Fujairah (top row) and Abu Dhabi samples (bottom row). Samples from adults (squares) and sperm (circles) tend to pair by colony identity. Samples from reciprocal larval crosses E7 x S8 and E8 x S7 (red triangles) are positioned midway between S7 and S8 along all plotted axes, suggesting equal contribution from both parents to their DNA methylation patterns. The sole egg sample, E8, is located close to S8 and A8, indicating that the transmission of epigenetic patterns from parent to either gamete type is unbiased.
Extended Data Fig. 3 Methylation trends against FST are origin-agnostic.
Genes were split into two groups, corresponding to genes with higher methylation in Abu Dhabi relative to Fujairah (red), and vice versa (blue). When correlated against genetic factors, both groups showed trends similar to Fig. 2, where methylation differences were calculated as absolute differences.
Extended Data Fig. 4 Outline of statistical tests performed.
Numbers denote biological replicates of different developmental stages and sample origins, while boxes denote the groupings used in the statistical tests. The initial GLM (green) tested whether both variables had significant interaction. Subsequently, the pair of t-tests (blue) identified genes that were differentially methylated across developmental stages, while the t-test (red) identified genes that were differentially methylated across sample origins.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Supplementary Dataset
Supplementary Data 1–8.
Rights and permissions
About this article
Cite this article
Liew, Y.J., Howells, E.J., Wang, X. et al. Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Chang. 10, 254–259 (2020). https://doi.org/10.1038/s41558-019-0687-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41558-019-0687-2
This article is cited by
-
Multi-omic characterization of mechanisms contributing to rapid phenotypic plasticity in the coral Acropora cervicornis under divergent environments
Coral Reefs (2024)
-
Stress memory in crops: what we have learned so far
Theoretical and Experimental Plant Physiology (2024)
-
Lineage-specific symbionts mediate differential coral responses to thermal stress
Microbiome (2023)
-
Associations between DNA methylation and gene regulation depend on chromatin accessibility during transgenerational plasticity
BMC Biology (2023)
-
Genome-wide DNA methylation patterns in bumble bee (Bombus vosnesenskii) populations from spatial-environmental range extremes
Scientific Reports (2023)