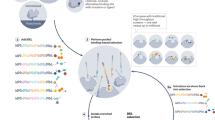
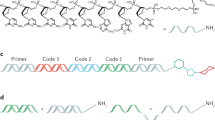
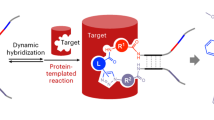
Abstract
DNA-encoded chemical libraries (DELs) have emerged as a powerful technology in drug discovery. The wide adoption of DELs in the pharmaceutical industry and the rapid advancements of DEL-compatible chemistry have further fuelled its development and applications. In general, a DEL has been considered as a massive binding assay to identify physical binders for individual protein targets. However, recent innovations demonstrate the capability of DELs to operate in the complex milieu of biological systems. In this Perspective, we discuss the recent progress in using DNA-encoded chemical libraries to interrogate complex biological targets and their potential to identify structures that elicit function or possess other useful properties. Future breakthroughs in these aspects are expected to catapult DEL to become a momentous technology platform not only for drug discovery but also to explore fundamental biology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
McCafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Wilson, D. S., Keefe, A. D. & Szostak, J. W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl Acad. Sci. USA 98, 3750–3755 (2001).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Hanes, J. & Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA 94, 4937–4942 (1997).
Neri, D. & Lerner, R. A. DNA-encoded chemical libraries: a selection system based on endowing organic compounds with amplifiable information. Annu. Rev. Biochem. 87, 479–502 (2018).
Patel, S., Badir, S. O. & Molander, G. A. Developments in photoredox-mediated alkylation for DNA-encoded libraries. Trends Chem. 3, 161–175 (2021).
Fitzgerald, P. R. & Paegel, B. M. DNA-encoded chemistry: drug discovery from a few good reactions. Chem. Rev. 121, 7155–7177 (2021).
Dickson, P. & Kodadek, T. Chemical composition of DNA-encoded libraries, past present and future. Org. Biomol. Chem. 17, 4676–4688 (2019).
Conole, D., J, H. H. & M, J. W. The maturation of DNA encoded libraries: opportunities for new users. Future Med. Chem. 13, 173–191 (2021).
Flood, D. T. et al. DNA encoded libraries: a visitor’s guide. Isr. J. Chem. 60, 268–280 (2020).
Götte, K., Chines, S. & Brunschweiger, A. Reaction development for DNA-encoded library technology: from evolution to revolution? Tetrahedron Lett. 61, 151889 (2020).
Lerner, R. A. & Brenner, S. DNA-encoded compound libraries as open source: a powerful pathway to new drugs. Angew. Chem. Int. Ed. 56, 1164–1165 (2017).
Brenner, S. & Lerner, R. A. Encoded combinatorial chemistry. Proc. Natl Acad. Sci. USA 89, 5381–5383 (1992).
Nielsen, J., Brenner, S. & Janda, K. D. Synthetic methods for the implementation of encoded combinatorial chemistry. J. Am. Chem. Soc. 115, 9812–9813 (1993).
Needels, M. C. et al. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc. Natl Acad. Sci. USA 90, 10700–10704 (1993).
Melkko, S., Scheuermann, J., Dumelin, C. E. & Neri, D. Encoded self-assembling chemical libraries. Nat. Biotechnol. 22, 568–574 (2004).
Gartner, Z. J. et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science 305, 1601–1605 (2004).
Halpin, D. R. & Harbury, P. B. DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2, 1015–1021 (2004).
Debaene, F., Mejias, L., Harris, J. L. & Winssinger, N. Synthesis of a PNA-encoded cysteine protease inhibitor library. Tetrahedron 60, 8677–8690 (2004).
Clark, M. A. et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654 (2009).
Song, M. & Hwang, G. T. DNA-encoded library screening as a core platform technology in drug discovery. Its synthetic method development and applications in DEL synthesis. J. Med. Chem. 63, 6578–6599 (2020).
Wichert, M. et al. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat. Chem. 7, 241–249 (2015).
Daguer, J. P. et al. DNA display of fragment pairs as a tool for the discovery of novel biologically active small molecules. Chem. Sci. 6, 739–744 (2015).
Daguer, J. P. et al. DNA-templated combinatorial assembly of small molecule fragments amenable to selection/amplification cycles. Chem. Sci. 2, 625–632 (2011).
Barluenga, S. et al. Novel PTP1B inhibitors identified by DNA display of fragment pairs. Bioorg. Med. Chem. Lett. 26, 1080–1085 (2016).
Reddavide, F. V., Lin, W., Lehnert, S. & Zhang, Y. DNA-encoded dynamic combinatorial chemical libraries. Angew. Chem. Int. Ed. 54, 7924–7928 (2015).
Reddavide, F. V. et al. Second generation DNA-encoded dynamic combinatorial chemical libraries. Chem. Commun. 55, 3753–3756 (2019).
Li, G. et al. Design, preparation, and selection of DNA-encoded dynamic libraries. Chem. Sci. 6, 7097–7104 (2015).
Zhou, Y. et al. DNA-encoded dynamic chemical library and its applications in ligand discovery. J. Am. Chem. Soc. 140, 15859–15867 (2018).
Deng, Y. et al. Selection of DNA-encoded dynamic chemical libraries for direct inhibitor discovery. Angew. Chem. Int. Ed. 59, 14965–14972 (2020).
Farrera-Soler, L. et al. PNA-based dynamic combinatorial libraries (PDCL) and screening of lectins. Bioorg. Med. Chem. 28, 115458 (2020).
Machida, T. et al. Dynamic cooperative glycan assembly blocks the binding of bacterial lectins to epithelial cells. Angew. Chem. Int. Ed. 56, 6762–6766 (2017).
Lenci, E., Baldini, L. & Trabocchi, A. Diversity-oriented synthesis as a tool to expand the chemical space of DNA-encoded libraries. Biorg. Med. Chem. 41, 116218 (2021).
Guasch, L., Reutlinger, M., Stoffler, D. & Wichert, M. Augmenting chemical space with DNA-encoded library technology and machine learning. Chimia 75, 105–107 (2021).
Martin, A., Nicolaou, C. A. & Toledo, M. A. Navigating the DNA encoded libraries chemical space. Commun. Chem. 3, 127 (2020).
McCloskey, K. et al. Machine learning on DNA-encoded libraries: a new paradigm for hit finding. J. Med. Chem. 63, 8857–8866 (2020).
Bobers, J. et al. Design of an automated reagent-dispensing system for reaction screening and validation with DNA-tagged substrates. ACS Comb. Sci. 22, 101–108 (2020).
Castanon, J. et al. Design and development of a technology platform for DNA-encoded library production and affinity selection. SLAS Discov. 23, 387–396 (2018).
MacConnell, A. B., McEnaney, P. J., Cavett, V. J. & Paegel, B. M. DNA-encoded solid-phase synthesis: encoding language design and complex oligomer library synthesis. ACS Comb. Sci. 17, 518–534 (2015).
Cochrane, W. G. et al. Activity-based DNA-encoded library screening. ACS Comb. Sci. 21, 425–435 (2019).
Hackler, A. L. et al. Off-DNA DNA-encoded library affinity screening. ACS Comb. Sci. 22, 25–34 (2020).
Shin, M. H., Lee, K. J. & Lim, H. S. DNA-encoded combinatorial library of macrocyclic peptoids. Bioconjug. Chem. 30, 2931–2938 (2019).
Mendes, K. R. et al. High-throughput identification of DNA-encoded IgG ligands that distinguish active and latent Mycobacterium tuberculosis infections. ACS Chem. Biol. 12, 234–243 (2017).
Arico-Muendel, C. C. From haystack to needle: finding value with DNA encoded library technology at GSK. MedChemComm 7, 1898–1909 (2016).
Belyanskaya, S. L. et al. Discovering drugs with DNA-encoded library technology: from concept to clinic with an inhibitor of soluble epoxide hydrolase. ChemBioChem 18, 837–842 (2017).
Harris, P. A. et al. Discovery of a first-in-class receptor Interacting protein 1 (RIP1) kinase specific clinical candidate (GSK2982772) for the treatment of inflammatory diseases. J. Med. Chem. 60, 1247–1261 (2017).
Cuozzo, J. W. et al. Novel autotaxin inhibitor for the treatment of idiopathic pulmonary fibrosis: a clinical candidate discovered using DNA-encoded chemistry. J. Med. Chem. 63, 7840–7856 (2020).
Ahn, S. et al. Allosteric ‘beta-blocker’ isolated from a DNA-encoded small molecule library. Proc. Natl Acad. Sci. USA 114, 1708–1713 (2017).
Ahn, S. et al. Small-molecule positive allosteric modulators of the beta2-adrenoceptor isolated from DNA-encoded libraries. Mol. Pharmacol. 94, 850–861 (2018).
Brown, D. G. et al. Agonists and antagonists of protease-activated receptor 2 discovered within a DNA-encoded chemical library using mutational stabilization of the target. SLAS Discov. 23, 429–436 (2018).
Figuerola-Conchas, A. et al. Small-molecule modulators of the ATPase VCP/p97 affect specific p97 cellular functions. ACS Chem. Biol. 15, 243–253 (2020).
Satz, A. L., Kuai, L. & Peng, X. Selections and screenings of DNA-encoded chemical libraries against enzyme and cellular targets. Bioorg. Med. Chem. Lett. 39, 127851 (2021).
Kodadek, T., Paciaroni, N. G., Balzarini, M. & Dickson, P. Beyond protein binding: recent advances in screening DNA-encoded libraries. Chem. Commun. 55, 13330–13341 (2019).
Goodnow R. A. Jr A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery (John Wiley & Sons, 2014).
Reiher, C. A., Schuman, D. P., Simmons, N. & Wolkenberg, S. E. Trends in hit-to-lead optimization following DNA-encoded library screens. ACS Med. Chem. Lett. 12, 343–350 (2021).
Shi, Y. et al. DNA-encoded libraries (DELs): a review of on-DNA chemistries and their output. RSC Adv. 11, 2359–2376 (2021).
Kunig, V. B. K., Potowski, M., Klika Skopic, M. & Brunschweiger, A. Scanning protein surfaces with DNA-encoded libraries. ChemMedChem 16, 1048–1062 (2021).
Madsen, D. et al. An overview of DNA-encoded libraries: a versatile tool for drug discovery. Prog. Med. Chem. 59, 181–249 (2020).
Gironda-Martínez, A., Donckele, E. J., Samain, F. & Neri, D. DNA-encoded chemical libraries: a comprehensive review with successful stories and future challenges. ACS Pharmacol. Trans. Sci. 4, 1265–1279 (2021).
Li, Y., Zimmermann, G., Scheuermann, J. & Neri, D. Quantitative PCR is a valuable tool to monitor the performance of DNA-encoded chemical library selections. ChemBioChem 18, 848–852 (2017).
Sannino, A. et al. Quantitative assessment of affinity selection performance by using DNA-encoded chemical libraries. ChemBioChem 20, 955–962 (2019).
Kim, D. et al. Application of a substrate-mediated selection with c-Src tyrosine kinase to a DNA-encoded chemical library. Molecules 24, 2764 (2019).
Denton, K. E. & Krusemark, C. J. Crosslinking of DNA-linked ligands to target proteins for enrichment from DNA-encoded libraries. MedChemComm 7, 2020–2027 (2016).
Sannino, A. et al. Critical evaluation of photo-cross-linking parameters for the implementation of efficient DNA-encoded chemical library selections. ACS Comb. Sci. 22, 204–212 (2020).
Chen, Q. et al. Exploring the lower limit of individual DNA-encoded library molecules in selection. SLAS Discov. 25, 523–529 (2020).
Andrade, H. et al. Using a PCR-based method to analyze and model large, heterogeneous populations of DNA. ChemBioChem 21, 1144–1149 (2020).
Satz, A. L. DNA encoded library selections and insights provided by computational simulations. ACS Chem. Biol. 10, 2237–2245 (2015).
Satz, A. L., Hochstrasser, R. & Petersen, A. C. Analysis of current DNA encoded library screening data indicates higher false negative rates for numerically larger libraries. ACS Comb. Sci. 19, 234–238 (2017).
McGregor, L. M., Jain, T. & Liu, D. R. Identification of ligand–target pairs from combined libraries of small molecules and unpurified protein targets in cell lysates. J. Am. Chem. Soc. 136, 3264–3270 (2014).
Chan, A. I., McGregor, L. M., Jain, T. & Liu, D. R. Discovery of a covalent kinase inhibitor from a DNA-encoded small-molecule library × protein library selection. J. Am. Chem. Soc. 139, 10192–10195 (2017).
Blakskjaer, P., Heitner, T. & Hansen, N. J. Fidelity by design: YoctoReactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr. Opin. Chem. Biol. 26, 62–71 (2015).
Petersen, L. K. et al. Novel p38 alpha MAP kinase inhibitors identified from YoctoReactor DNA-encoded small molecule library. MedChemComm 7, 1332–1339 (2016).
Petersen, L. K. et al. Screening of DNA-encoded small molecule libraries inside a living cell. J. Am. Chem. Soc. 143, 2751–2756 (2021).
Zhao, P. et al. Selection of DNA-encoded small molecule libraries against unmodified and non-immobilized protein targets. Angew. Chem. Int. Ed. 53, 10056–10059 (2014).
Shi, B., Deng, Y. & Li, X. Polymerase-extension-based selection method for DNA-encoded chemical libraries against nonimmobilized protein targets. ACS Comb. Sci. 21, 345–349 (2019).
Shi, B., Deng, Y., Zhao, P. & Li, X. Selecting a DNA-encoded chemical library against non-immobilized proteins using a ‘ligate–cross-link–purify’ strategy. Bioconjug. Chem. 28, 2293–2301 (2017).
Winssinger, N. & Harris, J. L. Microarray-based functional protein profiling using peptide nucleic acid-encoded libraries. Expert Rev. Proteomics 2, 937–947 (2005).
Harris, J. L. & Winssinger, N. PNA encoding (PNA = peptide nucleic acid): from solution-based libraries to organized microarrays. Chem. Eur. J. 11, 6792–6801 (2005).
Kochmann, S., Le, A. T. H., Hili, R. & Krylov, S. N. Predicting efficiency of NECEEM-based partitioning of protein binders from nonbinders in DNA-encoded libraries. Electrophoresis 39, 2991–2996 (2018).
Bao, J. et al. Predicting electrophoretic mobility of protein–ligand complexes for ligands from DNA-encoded libraries of small molecules. Anal. Chem. 88, 5498–5506 (2016).
Sprinz, K. I., Tagore, D. M. & Hamilton, A. D. Self-assembly of bivalent protein-binding agents based on oligonucleotide-linked organic fragments. Bioorg. Med. Chem. Lett. 15, 3908–3911 (2005).
Onda, Y. et al. A DNA-encoded chemical library based on peptide macrocycles. Chem. Eur. J. 27, 7160–7167 (2021).
Dal Corso, A. et al. Affinity enhancement of protein ligands by reversible covalent modification of neighboring lysine residues. Angew. Chem. Int. Ed. 57, 17178–17182 (2018).
Kollmann, C. S. et al. Application of encoded library technology (ELT) to a protein–protein interaction target: discovery of a potent class of integrin lymphocyte function-associated antigen 1 (LFA-1) antagonists. Bioorg. Med. Chem. 22, 2353–2365 (2014).
Richter, H. et al. DNA-encoded library-derived DDR1 inhibitor prevents fibrosis and renal function loss in a genetic mouse model of Alport syndrome. ACS Chem. Biol. 14, 37–49 (2019).
Xie, J. et al. Selection of small molecules that bind to and activate the insulin receptor from a DNA-encoded library of natural products. iScience 23, 101197 (2020).
Favalli, N. et al. Stereo- and regiodefined DNA-encoded chemical libraries enable efficient tumour-targeting applications. Nat. Chem. 13, 540–548 (2021).
Huang, Y. et al. Selection of DNA-encoded chemical libraries against endogenous membrane proteins on live cells. Nat. Chem. 13, 77–88 (2021).
Svensen, N., Diaz-Mochon, J. J. & Bradley, M. Decoding a PNA encoded peptide library by PCR: the discovery of new cell surface receptor ligands. Chem. Biol. 18, 1284–1289 (2011).
Svensen, N., Diaz-Mochon, J. J. & Bradley, M. Encoded peptide libraries and the discovery of new cell binding ligands. Chem. Commun. 47, 7638–7640 (2011).
Wu, Z. et al. Cell-based selection expands the utility of DNA-encoded small-molecule library technology to cell surface drug targets: identification of novel antagonists of the NK3 tachykinin receptor. ACS Comb. Sci. 17, 722–731 (2015).
Cai, B. et al. Selection of DNA-encoded libraries to protein targets within and on living cells. J. Am. Chem. Soc. 141, 17057–17061 (2019).
Li, G. et al. Photoaffinity labeling of small-molecule-binding proteins by DNA-templated chemistry. Angew. Chem. Int. Ed. 52, 9544–9549 (2013).
Schroeder, H. et al. Generation of live-cell microarrays by means of DNA-directed immobilization of specific cell-surface ligands. Angew. Chem. Int. Ed. 46, 4180–4183 (2007).
Stahl, S. et al. Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 35, 691–712 (2017).
Li, L. et al. Aptamer displacement reaction from live-cell surfaces and its applications. J. Am. Chem. Soc. 141, 17174–17179 (2019).
Oehler, S. et al. Affinity selections of DNA-encoded chemical libraries on carbonic anhydrase IX—expressing tumor cells reveal a dependence on ligand valence. Chem. Eur. J. 27, 8985–8993 (2021).
Litovchick, A. et al. Novel nucleic acid binding small molecules discovered using DNA-encoded chemistry. Molecules 24, 2026 (2019).
Blain, J. C. et al. Encoded libraries and methods of use for screening nucleic acid targets. World patent WO2019236644A1 (2019).
Zhang, J. et al. Identification of histone deacetylase (HDAC)-associated proteins with DNA-programmed affinity labeling. Angew. Chem. Int. Ed. 59, 17525–17532 (2020).
Komnatnyy, V. V., Nielsen, T. E. & Qvortrup, K. Bead-based screening in chemical biology and drug discovery. Chem. Commun. 54, 6759–6771 (2018).
Wu, L. et al. Aptamer-based detection of circulating targets for precision medicine. Chem. Rev. 121, 12035–12105 (2021).
Svensen, N. et al. Screening of a combinatorial homing peptide library for selective cellular delivery. Angew. Chem. Int. Ed. 50, 6133–6136 (2011).
Kolodny, G., Li, X. & Balk, S. Addressing cancer chemotherapeutic toxicity, resistance, and heterogeneity: novel theranostic use of DNA-encoded small molecule libraries. Bioessays 40, e1800057 (2018).
Cuozzo, J. W. et al. Discovery of a potent BTK inhibitor with a novel binding mode by using parallel selections with a DNA-encoded chemical library. ChemBioChem 18, 864–871 (2017).
Zambaldo, C. et al. Screening for covalent inhibitors using DNA-display of small molecule libraries functionalized with cysteine reactive moieties. MedChemComm 7, 1340–1351 (2016).
Zhu, Z. et al. Development of a selection method for discovering irreversible (covalent) binders from a DNA-encoded library. SLAS Discov 24, 169–174 (2018).
Zimmermann, G. et al. A specific and covalent JNK-1 ligand selected from an encoded self-assembling chemical library. Chem. Eur. J. 23, 8152–8155 (2017).
Daguer, J. P. et al. Identification of covalent bromodomain binders through DNA display of small molecules. Angew. Chem. Int. Ed. 54, 6057–6061 (2015).
Guilinger, J. P. et al. Novel irreversible covalent BTK inhibitors discovered using DNA-encoded chemistry. Bioorg. Med. Chem. 42, 116223 (2021).
Winssinger, N. et al. PNA-encoded protease substrate microarrays. Chem. Biol. 11, 1351–1360 (2004).
Diaz-Mochon, J. J., Bialy, L. & Bradley, M. Dual colour, microarray-based, analysis of 10,000 protease substrates. Chem. Commun. 14, 3984–3986 (2006).
Pouchain, D., Diaz-Mochon, J. J., Bialy, L. & Bradley, M. A 10,000 member PNA-encoded peptide library for profiling tyrosine kinases. ACS Chem. Biol. 2, 810–818 (2007).
Krusemark, C. J., Tilmans, N. P., Brown, P. O. & Harbury, P. B. Directed chemical evolution with an outsized genetic code. PLoS ONE 11, e0154765 (2016).
Jetson, R. R. & Krusemark, C. J. Sensing enzymatic activity by exposure and selection of DNA-encoded probes. Angew. Chem. Int. Ed 55, 9562–9566 (2016).
Roy, A., Koesema, E. & Kodadek, T. J. High-throughput quality control assay for the solid-phase synthesis of DNA-encoded libraries of macrocycles. Angew. Chem. Int. Ed. 60, 11983–11990 (2021).
MacConnell, A. B., Price, A. K. & Paegel, B. M. An integrated microfluidic processor for DNA-encoded combinatorial library functional screening. ACS Comb. Sci. 19, 181–192 (2017).
Borchardt, A. et al. Small molecule-dependent genetic selection in stochastic nanodroplets as a means of detecting protein–ligand interactions on a large scale. Chem. Biol. 4, 961–968 (1997).
Schapira, M., Calabrese, M. F., Bullock, A. N. & Crews, C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 (2019).
Zhu, Z. et al. Design and application of a DNA-encoded macrocyclic peptide library. ACS Chem. Biol. 13, 53–59 (2018).
Disch, J. S. et al. Bispecific estrogen receptor alpha degraders incorporating novel binders identified using DNA-encoded chemical library screening. J. Med. Chem. 64, 5049–5066 (2021).
Andersen, J, N. et al. Degradation of immuno-oncology targets via proprietary PROTAC platform integrating DNA-encoded library technology and rational drug design. Cancer Res. 79, https://doi.org/10.1158/1538-7445.AM2019-1981 (2019).
Kanan, M. W. et al. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 431, 545–549 (2004).
Chen, Y., Kamlet, A. S., Steinman, J. B. & Liu, D. R. A biomolecule-compatible visible-light-induced azide reduction from a DNA-encoded reaction-discovery system. Nat. Chem. 3, 146–153 (2011).
Krattiger, P., McCarthy, C., Pfaltz, A. & Wennemers, H. Catalyst-substrate coimmobilization: A strategy for catalysts discovery in split-and-mix libraries. Angew. Chem. Int. Ed. 42, 1722–1724 (2003).
Hook, K. D., Chambers, J. T. & Hili, R. A platform for high-throughput screening of DNA-encoded catalyst libraries in organic solvents. Chem. Sci. 8, 7072–7076 (2017).
Brudno, Y., Birnbaum, M. E., Kleiner, R. E. & Liu, D. R. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat. Chem. Biol. 6, 148–155 (2010).
Hili, R., Niu, J. & Liu, D. R. DNA ligase-mediated translation of DNA into densely functionalized nucleic acid polymers. J. Am. Chem. Soc. 135, 98–101 (2013).
Kong, D., Yeung, W. & Hili, R. In vitro selection of diversely functionalized aptamers. J. Am. Chem. Soc. 139, 13977–13980 (2017).
Yeldell, S. B. & Seitz, O. Nucleic acid constructs for the interrogation of multivalent protein interactions. Chem. Soc. Rev. 49, 6848–6865 (2020).
Barluenga, S. & Winssinger, N. PNA as a biosupramolecular tag for programmable assemblies and reactions. Acc. Chem. Res. 48, 1319–1331 (2015).
Flood, D. T. et al. Expanding reactivity in DNA-encoded library synthesis via reversible binding of DNA to an inert quaternary ammonium support. J. Am. Chem. Soc. 141, 9998–10006 (2019).
Skopic, M. K. et al. Micellar Brønsted acid mediated synthesis of DNA-tagged heterocycles. J. Am. Chem. Soc. 141, 10546–10555 (2019).
Hunter, J. H. et al. Highly efficient on-DNA amide couplings promoted by micelle forming surfactants for the synthesis of DNA encoded libraries. Chem. Sci. 12, 9475–9484 (2021).
Skopic, M. K. et al. Acid- and Au(I)-mediated synthesis of hexathymidine-DNA-heterocycle chimeras, an efficient entry to DNA-encoded libraries inspired by drug structures. Chem. Sci. 8, 3356–3361 (2017).
Potowski, M. et al. Chemically stabilized DNA barcodes for DNA-encoded chemistry. Angew. Chem. Int. Ed. 60, 19744–19749 (2021).
Rama-Garda, R. et al. Normalization of DNA encoded library affinity selection results driven by high throughput sequencing and HPLC purification. Bioorg. Med. Chem. 40, 116178 (2021).
Komar, P. & Kalinic, M. Denoising DNA encoded library screens with sparse learning. ACS Comb. Sci. 22, 410–421 (2020).
Gerry, C. J., Wawer, M. J., Clemons, P. A. & Schreiber, S. L. DNA barcoding a complete matrix of stereoisomeric small molecules. J. Am. Chem. Soc. 141, 10225–10235 (2019).
Faver, J. C. et al. Quantitative comparison of enrichment from DNA-encoded chemical library selections. ACS Comb. Sci. 21, 75–82 (2019).
Kuai, L., O’Keeffe, T. & Arico-Muendel, C. Randomness in DNA encoded library selection data can be modeled for more reliable enrichment calculation. SLAS Discov. 23, 405–416 (2018).
Denton, K. E. et al. Robustness of in vitro selection assays of DNA-encoded peptidomimetic ligands to CBX7 and CBX8. SLAS Discov. 23, 417–428 (2018).
Su, W. et al. Triaging of DNA-encoded library selection results by high-throughput resynthesis of DNA-conjugate and affinity selection mass spectrometry. Bioconjug. Chem. 32, 1001–1007 (2021).
Prudent, R. et al. Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat. Rev. Chem. 5, 62–71 (2021).
Kielar, C. et al. Pharmacophore nanoarrays on DNA origami substrates as a single-molecule assay for fragment-based drug discovery. Angew. Chem. Int. Ed. 57, 14873–14877 (2018).
Acknowledgements
This work was supported by grants from the Research Grants Council of the Hong Kong Special Administrative Region, China (AoE/P-705/16, 17301118, 17111319, 17303220, 17300321 and C7005-20G), and from the National Natural Science Foundation of China (21572014, 21877093, 21907011 and 91953119), the Fundamental Research Funds for the Central Universities (project nos. 2020CQJQY-Z002 and 2021CDJYGRH-002) and Chongqing Research and Frontier Technology (cstc2020jcyj-jqX0009 and cstc2021jcyj-cxttX0002). We also acknowledge the funding support from the Laboratory for Synthetic Chemistry and Chemical Biology under the Health@InnoHK Program launched by the Innovation and Technology Commission, The Government of Hong Kong Special Administrative Region of the People’s Republic of China.
Author information
Authors and Affiliations
Contributions
Y.H., Y.L. and X.L. contributed to the discussions and co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Nicolas Winssinger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, Y., Li, Y. & Li, X. Strategies for developing DNA-encoded libraries beyond binding assays. Nat. Chem. 14, 129–140 (2022). https://doi.org/10.1038/s41557-021-00877-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00877-x
This article is cited by
-
Protein-templated ligand discovery via the selection of DNA-encoded dynamic libraries
Nature Chemistry (2024)
-
Small-molecule discovery through DNA-encoded libraries
Nature Reviews Drug Discovery (2023)