Abstract
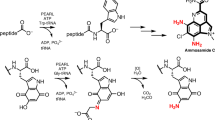
Natural products with rare functional groups are likely to be constructed by unique biosynthetic enzymes. One such rare functional group is the O-methyl nitronate, which can undergo [3 + 2] cycloaddition reactions with olefins in mild conditions. O-methyl nitronates are found in some natural products; however, how such O-methyl nitronates are assembled biosynthetically is unknown. Here we show that the assembly of the O-methyl nitronate in the natural product enteromycin carboxamide occurs via activation of glycine on a peptidyl carrier protein, followed by reaction with a diiron oxygenase to give a nitronate intermediate and then with a methyltransferase to give an O-methyl nitronate. Guided by the discovery of this pathway, we then identify related cryptic biosynthetic gene cassettes in other bacteria and show that these alternative gene cassettes can, instead, facilitate oxidative denitrification of glycine-derived nitronates. Altogether, our work reveals bifurcating pathways from a central glycine-derived nitronate intermediate in bacteria.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The nucleotide sequence of the etm gene cluster is deposited at NCBI (accession no. MW367897). All other data described are provided in this Article and the associated Supplementary Information.
References
Kornblum, N. & Brown, R. A. The synthesis and characterization of nitronic esters. J. Am. Chem. Soc. 86, 2681–2687 (1964).
Denmark, S. E. & Thorarensen, A. Tandem [4 + 2]/[3 + 2] cycloadditions of nitroalkenes. Chem. Rev. 96, 137–166 (1996).
Denmark, S. E. & Montgomery, J. I. Synthesis of cis,cis,cis,cis-[5.5.5.4]-1-azafenestrane with discovery of an unexpected dyotropic rearrangement. Angew. Chem. Int. Ed. 44, 3732–3736 (2005).
Mitscher, L. A., McCrae, W. & DeVoe, S. E. The structural characterization of enteromycin carboxamide. Tetrahedron 21, 267–271 (1965).
Herr, R., Argoudelis, A., Bergy, M. & Jahnke, H. Three chemically related metabolites of Streptomyces. I. Isolation and characterization. Antimicrob. Agents Chemother. 10, 100–104 (1964).
Wiley, P. F., Herr, R. R., MacKellar, F. A. & Argoudelis, A. D. Three chemically related metabolites of Streptomyces. II. Structural studies. J. Org. Chem. 30, 2330–2334 (1965).
Kanemasa, S., Kaga, S. & Wada, E. Stability of electron deficient activated nitronates under neutral and Lewis acid catalysed conditions. Facile nitronate cycloaddition reactions to the magnesium alkoxides of allylic alcohols leading to isoxazolidines and isoxazolines. Tetrahedron Lett. 39, 8865–8868 (1998).
He, H.-Y., Henderson, A. C., Du, Y.-L. & Ryan, K. S. Two-enzyme pathway links l-arginine to nitric oxide in N-nitroso biosynthesis. J. Am. Chem. Soc. 141, 4026–4033 (2019).
Mizuno, K. Structure of enteromycin. I. Derivatives and products in various reactions of enteromycin. Bull. Chem. Soc. Jpn 34, 1419–1425 (1961).
Nsikabaka, S., Harb, W. & Ruiz-López, M. F. The role of water on the acid-promoted E/Z isomerization of oximes in aqueous solution. J. Mol. Struc. Theochem. 764, 161–166 (2006).
Hollenhorst, M. A., Clardy, J. & Walsh, C. T. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl–dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48, 10467–10472 (2009).
Morlot, C. et al. Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae. Nat. Commun. 9, 3180 (2018).
Scofield, M. A., Lewis, W. S. & Schuster, S. Nucleotide sequence of Escherichia coli asnB and deduced amino acid sequence of asparagine synthetase B. J. Biol. Chem. 265, 12895–12902 (1990).
Li, Y., Llewellyn, N. M., Giri, R., Huang, F. & Spencer, J. B. Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem. Biol. 12, 665–675 (2005).
Dunbar, K. L., Dell, M., Gude, F. & Hertweck, C. Reconstitution of polythioamide antibiotic backbone formation reveals unusual thiotemplated assembly strategy. Proc. Natl Acad. Sci. USA 117, 8850–8858 (2020).
Mocibob, M. et al. Homologues of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis. Proc. Natl Acad. Sci. USA 107, 14585–14590 (2010).
Winkler, R., Richter, M. E. A., Knüpfer, U., Merten, D. & Hertweck, C. Regio- and chemoselective enzymatic N-oxygenation in vivo, in vitro, and in flow. Angew. Chem. Int. Ed. 45, 8016–8018 (2006).
Choi, Y. S., Zhang, H., Brunzelle, J. S., Nair, S. K. & Zhao, H. In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc. Natl Acad. Sci. USA 105, 6858–6863 (2008).
Lu, H., Chanco, E. & Zhao, H. CmlI is an N-oxygenase in the biosynthesis of chloramphenicol. Tetrahedron 68, 7651–7654 (2012).
Graña, M. et al. The crystal structure of M. leprae ML2640c defines a large family of putative S-adenosylmethionine-dependent methyltransferases in mycobacteria. Protein Sci. 16, 1896–1904 (2007).
Sakata, T. et al. Origin of regioselectivity in the reactions of nitronate and enolate ambident anions. J. Org. Chem. 77, 10738–10744 (2012).
Arndt, F. & Rose, J. D. Relations between acidity and tautomerism. Part III. The nitro-group and the nitronic esters. J. Chem. Soc. (Resumed) 1935, 1–10 (1935).
Mizuno, K. Structure of enteromycin. IV. Bull. Chem. Soc. Jpn 34, 1633–1639 (1961).
Gadda, G. & Francis, K. Nitronate monooxygenase, a model for anionic flavin semiquinone intermediates in oxidative catalysis. Arch. Biochem. Biophys. 493, 53–61 (2010).
Ball, J., Salvi, F. & Gadda, G. Functional annotation of a presumed nitronate monooxygenase reveals a new class of NADH:quinone reductases. J. Biol. Chem. 291, 21160–21170 (2016).
Francis, K., Smitherman, C., Nishino, S. F., Spain, J. C. & Gadda, G. The biochemistry of the metabolic poison propionate 3-nitronate and its conjugate acid, 3-nitropropionate. IUBMB Life 65, 759–768 (2013).
Marroquin-Guzman, M. The Magnaporthe orysae nitrooxidative stress response suppresses rice innate immunity during blast disease. Nat. Microbiol. 2, 17054 (2017).
Fitzpatrick, P. F. Nitroalkane oxidase: structure and mechanism. Arch. Biochem. Biophys. 632, 41–46 (2017).
Li, Y. et al. Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes. Biochem Biophys. Res. Commun. 405, 344–348 (2011).
Zhang, J., Ma, W. & Tan, H. Cloning, expression and characterization of a gene encoding nitroalkane-oxidizing enzyme from Streptomyces ansochromogenes: a nitroalkane-oxidizing enzyme of Streptomyces. Eur. J. Biochem. 269, 6302–6307 (2002).
Bracher, P. J., Snyder, P. W., Bohall, B. R. & Whitesides, G. M. The relative rates of thiol–thioester exchange and hydrolysis for alkyl and aryl thioalkanoates in water. Origins Life Evol. B 41, 399–412 (2011).
Schaffer, J. E., Reck, M. R., Prasad, N. K. & Wencewicz, T. A. β-lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol. 13, 737–744 (2017).
Guo, Y.-Y. et al. Molecular mechanism of azoxy bond formation for azoxymycins biosynthesis. Nat. Commun. 10, 4420 (2019).
Kudo, K., Ozaki, T., Shin-ya, K., Nishiyama, M. & Kuzuyama, T. Biosynthetic origin of the hydroxamic acid moiety of trichostatin A: identification of unprecedented enzymatic machinery Involved in hydroxylamine transfer. J. Am. Chem. Soc. 139, 6799–6802 (2017).
Cortina, N. S., Revermann, O., Krug, D. & Müller, R. Identification and characterization of the althiomycin biosynthetic gene cluster in Myxococcus xanthus DK897. ChemBioChem 12, 1411–1416 (2011).
Kretsch, A. M. et al. Discovery of (dihydro)pyrazine N-oxides via genome mining in Pseudomonas. Org. Lett. 20, 4791–4795 (2018).
Jenul, C. et al. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 9, 1297 (2018).
Sieber, S. et al. Biosynthesis and structure activity relationship investigations of the diazeniumdiolate antifungal agent fragin. ChemBioChem 21, 1587–1592 (2020).
Morgan, G. L. & Li, B. In vitro reconstitution reveals a central role for the N-oxygenase PvfB in (dihydro)pyrazine-N-oxide and valdiazen biosynthesis. Angew. Chem. Int. Ed. 132, 21387–21391 (2020).
Patai, S. The Chemistry of Amino, Nitroso, and Nitro Compounds and Their Derivatives Vol. 29 (Wiley, 1982).
Job, V., Marcone, G. L., Pilone, M. S. & Pollegioni, L. Glycine oxidase from Bacillus subtilis. Characterization of a new flavoprotein. J. Biol. Chem. 277, 6985–6993 (2002).
Alves, A., Bassot, A., Bulteau, A.-L., Pirola, L. & Morio, B. Glycine metabolism and its alterations in obesity and metabolic diseases. Nutrients 11, 1356 (2019).
Zallot, R., Oberg, N. O. & Gerlt, J. A. ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol. 47, 77–85 (2018).
Krieg, P. A. & Melton, D. [25] In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 155, 397–415 (1987).
Bierman, M. et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992).
Du, Y.-L., Shen, X.-L., Yu, P., Bai, L.-Q. & Li, Y.-Q. Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl. Env. Microbiol. 77, 8415–8426 (2011).
Wang, M., Niikura, H., He, H.-Y., Daniel-Ivad, P. & Ryan, K. S. Biosynthesis of the N–N-bond-containing compound l-alanosine. Angew. Chem. Int. Ed. 59, 3881–3885 (2020).
Mason, D., Lummis, W. & Dietz, A. U-22956, a new antibiotic. I. Discovery and biological activity. Antimicrob. Agents Chemother. 10, 110 (1964).
Imai, H., Suzuki, K.-I., Takamura, S., Kadota, S. & Iwanami, M. A new enteromycin group antibiotic, YN-0165J-A produced by Streptomyces sp. J. Antibiot. 39, 601–602 (1986).
Takeuchi, M. et al. Nitracidomycins A and B, new enteromycin-group antibiotics. J. Antibiot. 42, 329–332 (1989).
Dose, B. et al. Food-poisoning bacteria employ a citrate synthase and a Type II NRPS to synthesize bolaamphiphilic lipopeptide antibiotics. Angew. Chem. Int. Ed. 59, 21535–21540 (2020).
Dashti, Y. et al. Discovery and biosynthesis of bolagladins: unusual lipodepsipeptides from Burkholderia gladioli clinical isolates. Angew. Chem. Int. Ed. 59, 21553–21561 (2020).
Zallot, R., Oberg, N. & Gerlt, J. A. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58, 4169–4182 (2019).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Choi, Y. S. et al. Cloning and heterologous expression of the spectinabilin biosynthetic gene cluster from Streptomyces spectabilis. Mol. Biosyst. 6, 336–338 (2010).
Scott, T. A., Heine, D., Qin, Z. & Wilkinson, B. An l-threonine transaldolase is required for l-threo-β-hydroxy-α-amino acid assembly during obafluorin biosynthesis. Nat. Commun. 8, 15935 (2017).
Tsutsumi, H. et al. Unprecedented cyclization catalysed by a cytochrome P450 in benzastatin biosynthesis. J. Am. Chem. Soc. 140, 6631–6639 (2018).
Wolf, F. et al. Characterization of the actinonin biosynthetic gene cluster. ChemBioChem 19, 1189–1195 (2018).
Leipoldt, F. et al. Warhead biosynthesis and the origin of structural diversity in hydroxamate metalloproteinase inhibitors. Nat. Commun. 8, 1965 (2017).
Acknowledgements
We thank the Natural Science and Engineering Research Council of Canada, the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research for financial support. We thank T. Huan and H. Chen for assistance with the mass spectrometry analysis, M. Ezhova for collecting the NMR data, A. Henderson for assistance on the bioinformatics analysis and P. Daniel‐Ivad and J. Hedges for feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
H.-Y.H. and K.S.R. designed the experiments, which H.-Y.H. performed. H.-Y.H. and K.S.R. analysed the data, and co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks Michelle Chang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 2 HPLC analysis of fermentation extracts of NRRL 3125 and NRRL 2697.
a, HPLC traces of extracts and purified compounds. I) Fermentation of Streptomyces achromogenes var. streptozoticus NRRL 3125, where we purified compounds E-2 and 1 by semi-preparative HPLC. II) Fermentation of Streptomyces achromogenes subsp. streptozoticus NRRL 2697, which is a previously reported producer of enteromycin and 25, although no enteromycin production was observed in these fermentation conditions in our laboratory. III) Purified Peak I (assigned as Z-2) analysed after several hours standing at room temperature, when some has converted to Peak II (assigned as E-2). IV) Purified E-2 run on HPLC after one week standing at RT, with minimal conversion to Peak I (assigned as Z-2). V) Purified Peak III (assigned as 1) analysed after 3 d standing at RT. b, Specific UV absorbance of peaks I, II, and III.
Extended Data Fig. 3 HPLC analysis of extracts of gene inactivation mutants.
Trace I, NRRL 3125 wild-type; Trace II, ΔetmG inactivation mutant; Trace III, ΔetmB inactivation mutant; Trace IV, ΔetmE inactivation mutant. Production of 1 and 2 is fully abolished in all three mutants.
Extended Data Fig. 4 Heat degradation of the product from the EtmE reaction.
a, Scheme of degradation and derivatization of formaldehyde with DNPH. b, EIC from LC-MS analysis of derivatized mixtures. m/z 198 and 210 were extracted, corresponding to DNPH and 10, respectively. I) Formaldehyde standard; II) Control reaction without EtmE; III) EtmE reaction. Negative mode was used for MS detection. 10 was detected in EtmE reaction, which confirmed the formation of O-methylnitronate in EtmE reaction.
Extended Data Fig. 5 Putative late steps in biosynthetic pathway of enteromycin-carboxamide (1) and U-15774 (2).
a, Biosynthetic gene cluster of 1. b, Putative late steps of the biosynthetic pathway. We propose that condensation of Gly-derived O-methyl nitronate 9 and Asp-derived β-aminopropionate 5, catalysed by putative 3-oxoacyl-[acyl-carrier-protein] synthase EtmF, gives a dipeptide intermediate, which is further modified via dehydrogenation (by putative dehydrogenases EtmK or EtmL), thioester hydrolysis (by putative thioesterase EtmQ) and transamination (by putative asparagine synthetase EtmT) to form 1. Remarkably, dehydrogenation may occur on 5 before condensation, as displayed in Fig. 1d of main text. A very recent study on the biosynthetic pathway to bolagladins shows that an enzyme related to EtmL, called BolQ, desaturates a β-aminopropionate unit, suggesting EtmL could have a similar function in 1 biosynthesis51,52. Additional genes in the enteromycin gene cluster include etmC and etmM, which encode putative 4'- phosphopantetheinyl transferases, which we propose activate two peptidyl carrier proteins EtmD and EtmI, respectively. The gene etmN encodes a putative S-adenosylmethionine synthetase, which might catalyse the formation of SAM for SAM-dependent methyltransferase EtmE. For the biosynthesis of oxime 2, one scenario is that it derives from a nitroso intermediate 7, detected in the EtmA reaction, which reacts with 5 directly catalysed by EtmF. However, the lack of 2 from ΔetmE (Extended Data Fig. 3) conflicts with this scenario. Previous studies revealed that 1 can convert to 2 under heating. We observed that purified 1 decomposed slowly to E- and Z-2 even at room temperature (Extended Data Fig. 2). Those results suggest a possible alternate nonenzymatic pathway to generate 2 from 1 in vivo.
Extended Data Fig. 6 Reaction of NMO Orf1480 with carrier protein-tethered nitronates.
a, Scheme of enzymatic reactions and chemical derivatization using DNPH. The nitroacetyl-S-carrier protein was generated from EtmA or UnkA reactions with unlabeled or labeled Gly as the substrate, as described for each panel. b,c, LC-MS analysis of derivatized mixtures in negative ion mode. b, Extracted-ion chromatogram (EIC) from LC-MS analysis, with m/z 198, 253 and 254 extracted, corresponding to DNPH, 15 and 15’, respectively. I) Derivatization of a glyoxylate chemical standard; II) Derivatization of the reaction with unlabeled Gly as the substrate; III) Derivatization of the reaction with 2-13C-15N-Gly as the substrate. c, MS signals displayed by each peak. I) Derivatization of the reaction with unlabeled Gly as the substrate; II) Derivatization of the reaction with 2-13C-15N-Gly as the substrate. Two peaks, consistent with DNPH-derivatized glyoxylate 15, were observed, which are assigned as isomers of carbon-nitrogen double bond (E-/Z-15), although which peak is E- or Z-form is unknown. As expected, comparing with the results of unlabeled-Gly added mixture, we detected +1 Da peak in the mixture of 2-13C-15N-Gly added, which is consistent with loss of one nitrogen and confirmed the formation of nitronate.
Extended Data Fig. 7 Sequence-similarity network of Pfam 11583.
A sequence-similarity network was generated using the EFI-Enzyme Similarity Tool, which analyses all proteins in UniProt53. Edges connect sequences that align with an E-value threshold of 1.0 x 10−36. Sequences with identities exceeding 90 % are represented by a single node. The network was visualized by Cytoscape54 using prefuse force directed openCL layout. Four identified clusters labeled are listed that contain known biosynthetic enzymes AurF17,18, SpnF55, CmlI19, ObiL (ObaC)32,56, AzoC33, BezJ57, ActI58, MatA59, TsnB734, HamC38, PvfB36,39 and AlmD35, all of them belonging to Pfam 11583.
Extended Data Fig. 8 Reactions catalysed by known biosynthetic enzymes belonging to Pfam 11583.
The schemes show characterized reactions of members of Pfam 11583. Not shown are two CmlI and ObiL (ObaC), which catalyse similar reactions to AurF in in the biosynthesis of chloramphenicol19 and obafluorin32,56, respectively. Note that ActI and MatA have not yet been experimentally characterized, and putative reactions are shown58,59.
Supplementary information
Supplementary Information
Chemical synthesis and Supplementary Tables 1–5, Figs. 1–20 and NMR spectra.
Rights and permissions
About this article
Cite this article
He, HY., Ryan, K.S. Glycine-derived nitronates bifurcate to O-methylation or denitrification in bacteria. Nat. Chem. 13, 599–606 (2021). https://doi.org/10.1038/s41557-021-00656-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00656-8
This article is cited by
-
Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis
Nature Chemical Biology (2024)