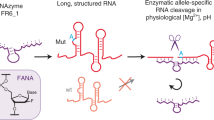
Abstract
Efforts to use RNA-cleaving DNA enzymes (DNAzymes) as gene-silencing agents in therapeutic applications have stalled due to their low efficacy in clinical trials. Here we report a xeno-nucleic-acid-modified version of the classic DNAzyme 10–23 that achieves multiple-turnover activity under cellular conditions and resists nuclease digestion. The new reagent, X10–23, overcomes the problem of product inhibition, which limited previous 10–23 designs, using molecular chemotypes with DNA, 2′-fluoroarabino nucleic acid and α-l-threofuranosyl nucleic acid backbone architectures that balance the effects of enhanced biological stability with RNA hybridization and divalent metal ion coordination. In cultured mammalian cells, X10–23 facilitates persistent gene silencing by efficiently degrading exogenous and endogenous messenger RNA transcripts. Together, these results demonstrate that new molecular chemotypes can improve the activity and stability of DNAzymes, and may provide a new route for nucleic acid enzymes to reach the clinic.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files. Source data are provided with this paper.
References
Santoro, S. W. & Joyce, G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA 94, 4262–4266 (1997).
Joyce, G. F. RNA cleavage by the 10–23 DNA enzyme. Methods Enzymol. 341, 503–517 (2001).
Khvorova, A. & Watts, J. K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35, 238–248 (2017).
Santoro, S. W. & Joyce, G. F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37, 13330–13342 (1998).
Appaiahgari, M. B. & Vrati, S. DNAzyme-mediated inhibition of Japanese encephalitis virus replication in mouse brain. Mol. Ther. 15, 1593–1599 (2007).
Xie, Y. Y. et al. Inhibition of respiratory syncytial virus in cultured cells by nucleocapsid gene targeted deoxyribozyme (DNAzyme). Antivir. Res. 71, 31–41 (2006).
Schubert, S. et al. RNA cleaving ‘10–23′ DNAzymes with enhanced stability and activity. Nucleic Acids Res. 31, 5982–5992 (2003).
Fokina, A. A., Meschaninova, M. I., Durfort, T., Venyaminova, A. G. & Francois, J. C. Targeting insulin-like growth factor I with 10–23 DNAzymes: 2′-O-methyl modifications in the catalytic core enhance mRNA cleavage. Biochemistry 51, 2181–2191 (2012).
Takahashi, H. et al. A new modified DNA enzyme that targets influenza virus A mRNA inhibits viral infection in cultured cells. FEBS Lett. 560, 69–74 (2004).
Vester, B. et al. LNAzymes: incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J. Am. Chem. Soc. 124, 13682–13683 (2002).
Fokina, A. A., Chelobanov, B. P., Fujii, M. & Stetsenko, D. A. Delivery of therapeutic RNA-cleaving oligodeoxyribonucleotides (deoxyribozymes): from cell culture studies to clinical trials. Expert. Opin. Drug Deliv. 14, 1077–1089 (2017).
Cho, E. A. et al. Safety and tolerability of an intratumorally injected DNAzyme, Dz13, in patients with nodular basal-cell carcinoma: a phase 1 first-in-human trial (DISCOVER). Lancet 381, 1835–1843 (2013).
Cao, Y. et al. Therapeutic evaluation of Epstein–Barr virus-encoded latent membrane protein-1 targeted DNAzyme for treating of nasopharyngeal carcinomas. Mol. Ther. 22, 371–377 (2014).
Hohlfeld, J. et al. Safety profile and pharmacokinetics of an inhaled GATA-3-specific DNAzyme in a phase Ib study in patients with stable allergic asthma. Eur. Respir. J. 42, 4856 (2013).
Cai, H. et al. DNAzyme targeting c-jun suppresses skin cancer growth. Sci. Transl. Med. 4, 139ra182 (2012).
Dicke, T. et al. Absence of unspecific innate immune cell activation by GATA-3-specific DNAzymes. Nucleic Acid Ther. 22, 117–126 (2012).
Chaput, J. C., Herdewijn, P. & Hollenstein, M. Orthogonal genetic systems. ChemBioChem 21, 1408–1411 (2020).
Damha, M. J. et al. Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates for ribonuclease H. J. Am. Chem. Soc. 120, 12976–12977 (1998).
Wilds, C. J. & Damha, M. J. 2′-Deoxy-2′-fluoro-β-d-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 28, 3625–3635 (2000).
Jakobsen, M. R., Haasnoot, J., Wengel, J., Berkhout, B. & Kjems, J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 4, 29 (2007).
Wolf, F. I., Torsello, A., Fasanella, S. & Cittadini, A. Cell physiology of magnesium. Mol. Aspects Med. 24, 11–26 (2003).
Jung, D. W., Apel, L. & Brierley, G. P. Matrix free Mg2+ changes with metabolic state in isolated heart mitochondria. Biochemistry 29, 4121–4128 (1990).
Cozens, C. et al. Enzymatic synthesis of nucleic acids with defined regioisomeric 2′–5′ linkages. Angew. Chem. Int. Ed. 54, 15570–15573 (2015).
Schöning, K. U. et al. Chemical etiology of nucleic acid structure: the ɑ-threofuranosyl-(3′→2′) oligonucleotide system. Science 290, 1347–1351 (2000).
Culbertson, M. C. et al. Evaluating TNA stability under simulated physiological conditions. Bioorg. Med. Chem. Lett. 26, 2418–2421 (2016).
Barrett, S. E. et al. An in vivo evaluation of amphiphilic, biodegradable peptide copolymers as siRNA delivery agents. Int. J. Pharm. 466, 58–67 (2014).
Cummins, L. L. et al. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23, 2019–2024 (1995).
Prakash, T. P. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem. Biodivers. 8, 1616–1641 (2011).
Farber, S., D’Angio, G., Evans, A. & Mitus, A. Clinical studies on actinomycin D with special reference to Wilms’ tumor in children. Ann. NY Acad. Sci. 89, 421–425 (1960).
Bensaude, O. Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011).
Chiosea, S. I., Sherer, C. K., Jelic, T. & Dacic, S. KRAS mutant allele-specific imbalance in lung adenocarcinoma. Mod. Pathol. 24, 1571–1577 (2011).
Krasinskas, A. M., Moser, A. J., Saka, B., Adsay, N. V. & Chiosea, S. I. KRAS mutant allele-specific imbalance is associated with worse prognosis in pancreatic cancer and progression to undifferentiated carcinoma of the pancreas. Mod. Pathol. 26, 1346–1354 (2013).
Hartman, D. J., Davison, J. M., Foxwell, T. J., Nikiforova, M. N. & Chiosea, S. I. Mutant allele-specific imbalance modulates prognostic impact of KRAS mutations in colorectal adenocarcinoma and is associated with worse overall survival. Int. J. Cancer 131, 1810–1817 (2012).
McCormick, F. KRAS as a therapeutic target. Clin. Cancer Res. 21, 1797–1801 (2015).
Young, D. D., Lively, M. O. & Deiters, A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 132, 6183–6193 (2010).
Fokina, A. A., Stetsenko, D. A. & Francois, J. C. DNA enzymes as potential therapeutics: towards clinical application of 10–23 DNAzymes. Expert Opin. Biol. Ther. 15, 689–711 (2015).
Hollenstein, M., Hipolito, C. J., Lam, C. H. & Perrin, D. M. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb. Sci. 15, 174–182 (2013).
Wang, Y., Liu, E., Lam, C. H. & Perrin, D. M. A densely modified M(2+)-independent DNAzyme that cleaves RNA efficiently with multiple catalytic turnover. Chem. Sci. 9, 1813–1821 (2018).
Silverman, S. K. Catalytic DNA: scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 41, 595–609 (2016).
Chaput, J. C., Yu, H. & Zhang, S. The emerging world of synthetic genetics. Chem. Biol. 19, 1360–1371 (2012).
Pinheiro, V. B., Loakes, D. & Holliger, P. Synthetic polymers and their potential as genetic materials. Bioessays 35, 113–122 (2012).
Anosova, I. et al. The structural diversity of artificial genetic polymers. Nucleic Acids Res. 44, 1007–1021 (2016).
Kim, J. et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 381, 1644–1652 (2019).
Liao, H. et al. Development of allele-specific therapeutic siRNA in Meesmann epithelial corneal dystrophy. PLoS ONE 6, e28582 (2011).
Christie, K. A. et al. Towards personalised allele-specific CRISPR gene editing to treat autosomal dominant disorders. Sci. Rep. 7, 16174 (2017).
Sau, S. P., Fahmi, N. E., Liao, J.-Y., Bala, S. & Chaput, J. C. A scalable synthesis of α-l-threose nucleic acid monomers. J. Org. Chem. 81, 2302–2307 (2016).
Acknowledgements
This work was supported by the W. M. Keck Foundation. Y.W. was supported by a postdoctoral fellowship from the Simons Collaboration on the Origins of Life.
Author information
Authors and Affiliations
Contributions
J.C.C. and R.C.S. conceived the project and designed the experiments. Y.W. and K.N. performed the experiments. J.C.C. wrote the manuscript with drafts from Y.W. and K.N. All the authors reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors and the University of California-Irvine have filed a patent application on the X10–23 reagent.
Additional information
Peer review information Nature Chemistry thanks Yingfu Li, Chuanzheng Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mechanistic analysis of X10-23.
Representative gels showing RNA cleavage activity in the presence and absence of RNase H for an internal segment of GFP (a-c) and the first exon segment of KRAS RNA (d-f). Color code: RNA (red), DNA (black), FANA (orange), and TNA (blue). (a,d) X10-23 with an active catalytic core. (b,e) X10-23 with an inactive catalytic core. (c,f) X10-23 with an active catalytic core that does not hybridize to the RNA target. All assays were performed in buffer containing 0.5 mM MgCl2 and 150 mM NaCl at 37 °C (pH 7.5) with 1 μM substrate and 1 μM enzyme. Nuclease reactions included 0.1 unit/μL of RNase H. S: full-length substrate, P: 5’ cleavage product. Molecular weight markers indicated to the right of the gel.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1 and 2, and Source Data.
Source data
Source Data Fig. 2
Uncropped gel from Fig. 2.
Source Data Fig. 3
Uncropped gels from Fig. 3a.
Source Data Fig. 4
Uncropped gels from Fig. 4c.
Source Data Extended Data Fig. 1
Uncropped gels from Extended Data Fig. 1.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Nguyen, K., Spitale, R.C. et al. A biologically stable DNAzyme that efficiently silences gene expression in cells. Nat. Chem. 13, 319–326 (2021). https://doi.org/10.1038/s41557-021-00645-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00645-x
This article is cited by
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Nature Communications (2023)
-
Simultaneous identification of viruses and viral variants with programmable DNA nanobait
Nature Nanotechnology (2023)
-
Chemical evolution of an autonomous DNAzyme with allele-specific gene silencing activity
Nature Communications (2023)
-
Discovering covalent inhibitors of protein–protein interactions from trillions of sulfur(VI) fluoride exchange-modified oligonucleotides
Nature Chemistry (2023)
-
An autocatalytic multicomponent DNAzyme nanomachine for tumor-specific photothermal therapy sensitization in pancreatic cancer
Nature Communications (2023)