Abstract
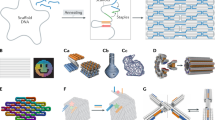
DNA origami has emerged as a highly programmable method to construct customized objects and functional devices in the 10–100 nm scale. Scaling up the size of the DNA origami would enable many potential applications, which include metamaterial construction and surface-based biophysical assays. Here we demonstrate that a six-helix bundle DNA origami nanostructure in the submicrometre scale (meta-DNA) could be used as a magnified analogue of single-stranded DNA, and that two meta-DNAs that contain complementary ‘meta-base pairs’ can form double helices with programmed handedness and helical pitches. By mimicking the molecular behaviours of DNA strands and their assembly strategies, we used meta-DNA building blocks to form diverse and complex structures on the micrometre scale. Using meta-DNA building blocks, we constructed a series of DNA architectures on a submicrometre-to-micrometre scale, which include meta-multi-arm junctions, three-dimensional (3D) polyhedrons, and various 2D/3D lattices. We also demonstrated a hierarchical strand-displacement reaction on meta-DNA to transfer the dynamic features of DNA into the meta-DNA. This meta-DNA self-assembly concept may transform the microscopic world of structural DNA nanotechnology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the data are available within this paper and its Supplementary Information. All the data are also available from the corresponding authors upon request.
Code availability
The code used for the ox-DNA simulation is available from the corresponding authors upon request.
Change history
24 November 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41557-020-00611-z
References
Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Liu, X. et al. Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Zhang, H. et al. Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 48, 6405–6407 (2012).
Marchi, A. N., Saaem, I., Vogen, B. N., Brown, S. & LaBean, T. H. Toward larger DNA origami. Nano Lett. 14, 5740–5747 (2014).
Zhao, Z., Yan, H. & Liu, Y. A route to scale up DNA origami using DNA tiles as folding staples. Angew. Chem. Int. Ed. 49, 1414–1417 (2010).
Zhao, Z., Liu, Y. & Yan, H. Organizing DNA origami tiles into larger structures using preformed scaffold frames. Nano Lett. 11, 2997–3002 (2011).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 6, 763–772 (2011).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Liu, W., Zhong, H., Wang, R. & Seeman, N. C. Crystalline two-dimensional DNA-origami arrays. Angew. Chem. Int. Ed. 50, 264–267 (2011).
Wang, P. et al. Programming self-assembly of DNA origami honeycomb two-dimensional lattices and plasmonic metamaterials. J. Am. Chem. Soc. 138, 7733–7740 (2016).
Tikhomirov, G., Petersen, P. & Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 552, 67–71 (2017).
Wagenbauer, K. F., Sigl, C. & Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017).
Liedl, T., Högberg, B., Tytell, J., Ingber, D. E. & Shih, W. M. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 5, 520 (2010).
Kauert, D. J., Kurth, T., Liedl, T. & Seidel, R. Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett. 11, 5558–5563 (2011).
Lee, C., Kim, K. S., Kim, Y.-J., Lee, J. Y. & Kim, D.-N. Tailoring the mechanical stiffness of DNA nanostructures using engineered defects. ACS Nano 13, 8329–8336 (2019).
Zhang, D. Y. & Yurke, B. A. A DNA superstructure-based replicator without product inhibition. Nat. Comput. 5, 183–202 (2006).
Chandran, H., Gopalkrishnan, N., Yurke, B. & Reif, J. Meta-DNA: synthetic biology via DNA nanostructures and hybridization reactions. J. R. Soc. Interface 9, 1637–1653 (2012).
Chandran, H., Gopalkrishnan, N., Yurke, B. & Reif, J. in A Systems Theoretic Approach to Systems and Synthetic Biology II: Analysis and Design of Cellular Systems (eds Kulkarni, V., Stan, G. B. & Raman, K.) 171–200 (2014).
Woo, S. & Rothemund, P. W. K. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011).
Petersen, P., Tikhomirov, G. & Qian, L. Information-based autonomous reconfiguration in systems of interacting DNA nanostructures. Nat. Commun. 9, 5362 (2018).
Doye, J. P. et al. Coarse-graining DNA for simulations of DNA nanotechnology. Phys. Chem. Chem. Phys. 15, 20395–20414 (2013).
Studdert, D. S., Patroni, M. & Davis, R. C. Circular dichroism of DNA: temperature and salt dependence. Biopolymers 11, 761–779 (1972).
Siavashpouri, M. et al. Molecular engineering of chiral colloidal liquid crystals using DNA origami. Nat. Mater. 16, 849–856 (2017).
Liu, Y., Ke, Y. & Yan, H. Self-assembly of symmetric finite-size DNA nanoarrays. J. Am. Chem. Soc. 127, 17140–17141 (2005).
Han, D. et al. Single-stranded DNA and RNA origami. Science 358, eaao2648 (2017).
Nguyen, L., Döblinger, M., Liedl, T. & Heuer-Jungemann, A. DNA-origami-templated silica growth by sol–gel chemistry. Angew. Chem. Int. Ed. 58, 912–916 (2019).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Wei, B., Dai, M. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Kallenbach, N. R., Ma, R.-I. & Seeman, N. C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 305, 829–831 (1983).
Fu, T. J. & Seeman, N. C. DNA double-crossover molecules. Biochemistry 32, 3211–3220 (1993).
Geary, C., Rothemund, P. W. & Andersen, E. S. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 345, 799–804 (2014).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Kilchherr, F. et al. Single-molecule dissection of stacking forces in DNA. Science 353, aaf5508 (2016).
Zhou, T. et al. pH-responsive size-tunable self-assembled DNA dendrimers. Angew. Chem. Int. Ed. 51, 11271–11274 (2012).
Yang, Y., Endo, M., Hidaka, K. & Sugiyama, H. Photo-controllable DNA origami nanostructures assembling into predesigned multiorientational patterns. J. Am. Chem. Soc. 134, 20645–20653 (2012).
Praetorius, F. et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017).
Qi, X. et al. Programming molecular topologies from single-stranded nucleic acids. Nat. Commun. 9, 4579 (2018).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
Thubagere, A. J. et al. A cargo-sorting DNA robot. Science 357, eaan6558 (2017).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Acknowledgements
The authors thank K. S. Kim, C. Lee and D.-N. Kim for the help of persistence length calculating. The authors thank D. Lowry for help with the TEM imaging and K. Lee for editing the paper. This work was financially supported by the US National Science Foundation, National Key R&D Program of China (2018YFA0902600), NSFC (21991134, 21834007, 21904060 and 21675167), the innovative research team of high-level local universities in Shanghai and the K. C. Wong Foundation at Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Contributions
G.Y., C.F. and H.Y. conceived the project. G.Y., T.P., F.W., S.J., L.L. and C.G. performed the research. H.L., E.P. and P.Š. performed the oxDNA simulation. X.J., X.L. and L.W. helped with the polyhedron silicification. G.Y., F.Z., Y.L., C.F. and H.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–100, Tables 1–80 and Sequences 1–6.
Supplementary Video 1
Dynamic animation of unwrapped ds-M-DNA of left-handed 660° design.
Supplementary Video 2
Dynamic animation of wrapped ds-M-DNA of left-handed 660° design.
Supplementary Video 3
Dynamic animation of unwrapped ds-M-DNA of right-handed 660° design.
Supplementary Video 4
Dynamic animation of wrapped ds-M-DNA of right-handed 660° design.
Rights and permissions
About this article
Cite this article
Yao, G., Zhang, F., Wang, F. et al. Meta-DNA structures. Nat. Chem. 12, 1067–1075 (2020). https://doi.org/10.1038/s41557-020-0539-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0539-8
This article is cited by
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Twisted moiré conductive thermal metasurface
Nature Communications (2024)
-
Biotemplated precise assembly approach toward ultra-scaled high-performance electronics
Nature Protocols (2023)
-
Multi-micron crisscross structures grown from DNA-origami slats
Nature Nanotechnology (2023)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)