Abstract
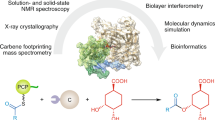
Type II polyketide synthases (PKSs) are multi-enzyme complexes that produce secondary metabolites of medical relevance. Chemical backbones of such polyketides are produced by minimal PKS systems that consist of a malonyl transacylase, an acyl carrier protein and an α/β heterodimeric ketosynthase. Here, we present X-ray structures of all ternary complexes that constitute the minimal PKS system for anthraquinone biosynthesis in Photorhabdus luminescens. In addition, we characterize this invariable core using molecular simulations, mutagenesis experiments and functional assays. We show that malonylation of the acyl carrier protein is accompanied by major structural rearrangements in the transacylase. Principles of an ongoing chain elongation are derived from the ternary complex with a hexaketide covalently linking the heterodimeric ketosynthase with the acyl carrier protein. Our results for the minimal PKS system provide mechanistic understanding of PKSs and a fundamental basis for engineering PKS pathways for future applications.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Crystallographic data have been deposited in the Protein Data Bank (https://www.rcsb.org) under the PDB IDs 6SM4, 6SM6, 6SMD, 6SMO and 6SMP. All other data are available in the text, methods, Supplementary Information, or upon request.
References
Hopwood, D. A. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97, 2465–2497 (1997).
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 48, 4688–4716 (2009).
McDaniel, R., Ebert-Khosla, S., Fu, H., Hopwood, D. A. & Khosla, C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc. Natl Acad. Sci. USA 91, 11542–11546 (1994).
Carreras, C. W., Gehring, A. M., Walsh, C. T. & Khosla, C. Utilization of enzymatically phosphopantetheinylated acyl carrier proteins and acetyl-acyl carrier proteins by the actinorhodin polyketide synthase. Biochemistry 36, 11757–11761 (1997).
Bao, W., Wendt-Pienkowski, E. & Hutchinson, C. R. Reconstitution of the iterative type II polyketide synthase for tetracenomycin F2 biosynthesis. Biochemistry 37, 8132–8138 (1998).
Chen, A., Re, R. N. & Burkart, M. D. Type II fatty acid and polyketide synthases: deciphering protein–protein and protein–substrate interactions. Nat. Prod. Rep. 35, 1029–1045 (2018).
Lambalot, R. H. & Walsh, C. T. Cloning, overproduction, and characterization of the Escherichia coli holo–acyl carrier protein synthase. J. Biol. Chem. 270, 24658–24661 (1995).
Serre, L., Verbree, E. C., Dauter, Z., Stuitje, A. R. & Derewenda, Z. S. The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5 Å resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270, 12961–12964 (1995).
Khosla, C., Gokhale, R. S., Jacobsen, J. R. & Cane, D. E. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68, 219–253 (1999).
Tsai, S.-C. The structural enzymology of iterative aromatic polyketide synthases: a critical comparison with fatty acid synthases. Annu. Rev. Biochem. 87, 503–531 (2018).
Sundaram, S. & Hertweck, C. On-line enzymatic tailoring of polyketides and peptides in thiotemplate systems. Curr. Opin. Chem. Biol. 31, 82–94 (2016).
Dutta, S. et al. Structure of a modular polyketide synthase. Nature 510, 512–517 (2014).
Li, X. et al. Structure–function analysis of the extended conformation of a polyketide synthase module. J. Am. Chem. Soc. 140, 6518–6521 (2018).
Hertweck, C., Luzhetskyy, A., Rebets, Y. & Bechthold, A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190 (2007).
Brachmann, A. O. et al. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. ChemBioChem 8, 1721–1728 (2007).
Zhou, Q. et al. Molecular mechanism of polyketide shortening in anthraquinone biosynthesis of Photorhabdus luminescens. Chem. Sci. 10, 6341–6349 (2019).
Cummings, M. et al. Assembling a plug-and-play production line for combinatorial biosynthesis of aromatic polyketides in Escherichia coli. PLoS Biol. 17, e3000347 (2019).
Walsh, C. T., Gehring, A. M., Weinreb, P. H., Quadri, L. E. & Flugel, R. S. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1, 309–315 (1997).
Dreier, J., Shah, A. N. & Khosla, C. Kinetic analysis of the actinorhodin aromatic polyketide synthase. J. Biol. Chem. 274, 25108–25112 (1999).
Fu, H., Hopwood, D. A. & Khosla, C. Engineered biosynthesis of novel polyketides: evidence for temporal, but not regiospecific, control of cyclization of an aromatic polyketide precursor. Chem. Biol. 1, 205–210 (1994).
Summers, R. G., Ali, A., Shen, B., Wessel, W. A. & Hutchinson, C. R. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry 34, 9389–9402 (1995).
Holak, T. A., Kearsley, S. K., Kim, Y. & Prestegard, J. H. Three-dimensional structure of acyl carrier protein determined by NMR pseudoenergy and distance geometry calculations. Biochemistry 27, 6135–6142 (1988).
Chan, D. I. & Vogel, H. J. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 430, 1–19 (2010).
Ollis, D. L. et al. The alpha/beta hydrolase fold. Protein Eng. 5, 197–211 (1992).
Pastore, A., Saudek, V., Ramponi, G. & Williams, R. J. P. Three-dimensional structure of acylphosphatase: refinement and structure analysis. J. Mol. Biol. 224, 427–440 (1992).
Oefner, C., Schulz, H., D’Arcy, A. & Dale, G. E. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D 62, 613–618 (2006).
Keatinge-Clay, A. T., Maltby, D. A., Medzihradszky, K. F., Khosla, C. & Stroud, R. M. An antibiotic factory caught in action. Nat. Struct. Mol. Biol. 11, 888–893 (2004).
Grammbitter, G. L. C. et al. An uncommon type II PKS catalyzes biosynthesis of aryl polyene pigments. J. Am. Chem. Soc. 141, 16615–16623 (2019).
Mathieu, M. et al. The 2.8 Å crystal structure of peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: a five-layered αβαβα structure constructed from two core domains of identical topology. Structure. 2, 797–808 (1994).
Robbins, T., Kapilivsky, J., Cane, D. E. & Khosla, C. Roles of conserved active site residues in the ketosynthase domain of an assembly line polyketide synthase. Biochemistry 55, 4476–4484 (2016).
Fu, H., Ebert-Khosla, S., Hopwood, D. A. & Khosla, C. Engineered biosynthesis of novel polyketides: dissection of the catalytic specificity of the act ketoreductase. J. Am. Chem. Soc. 116, 4166–4170 (1994).
Jakobi, K. & Hertweck, C. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J. Am. Chem. Soc. 126, 2298–2299 (2004).
Sherman, D. H., Kim, E. S., Bibb, M. J. & Hopwood, D. A. Functional replacement of genes for individual polyketide synthase components in Streptomyces coelicolor A3(2) by heterologous genes from a different polyketide pathway. J. Bacteriol. 174, 6184–6190 (1992).
Heath, R. J. & Rock, C. O. The Claisen condensation in biology. Nat. Prod. Rep. 19, 581–596 (2002).
Duchaud, E. et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–1313 (2003).
Kulak, N. A., Pichler, G., Paron, I., Nagaraj, N. & Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Roujeinikova, A. et al. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 365, 135–145 (2007).
Turk, D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr. D 69, 1342–1357 (2013).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Becke, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Lee, C., Yang, W. & Parr, R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988).
Schäfer, A., Huber, C. & Ahlrichs, R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 100, 5829–5835 (1994).
Grimme, S., Antony, J., Ehrlich, S. & Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H–Pu. J. Chem. Phys. 132, 154104 (2010).
Klamt, A. & Schüürmann, G. COSMO: a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc., Perkin Trans. 2, 799–805 (1993).
Huang, J. & MacKerell, A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Ahlrichs, R., Bär, M., Häser, M., Horn, H. & Kölmel, C. Electronic structure calculations on workstation computers: the program system turbomole. Chem. Phys. Lett. 162, 165–169 (1989).
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Riahi, S. & Rowley, C. N. The CHARMM-TURBOMOLE interface for efficient and accurate QM/MM molecular dynamics, free energies, and excited state properties. J. Comput. Chem. 35, 2076–2086 (2014).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Fenyö, D. et al. MALDI sample preparation: the ultra thin layer method. J. Vis. Exp. 192, 1–2 (2007).
Rühl, M. et al. Elucidation of chemical modifier reactivity towards peptides and proteins and the analysis of specific fragmentation by matrix-assisted laser desorption/ionization collision-induced dissociation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 33, 40–49 (2019).
Acknowledgements
We acknowledge financial support from the Deutsche Forschungsgemeinschaft (DFG) SPP 1617 (H.B.B.) and SFB 1035 (V.R.I.K.) projects, as well as from the LOEWE program of the state of Hesse as part of the MegaSyn research cluster (H.B.B.). We thank the staff of beamline X06SA at the Paul Scherrer Institute (SLS, Villigen) for assistance during data collection, S. Fuchs for MALDI-MS analysis, and M. Karas for MALDI-MS access. The Swedish National Infrastructure for Computing (SNIC, 2019/2-3) provided computing time.
Author information
Authors and Affiliations
Contributions
H.B.B. and M.G. initiated and supervised the project; Q.Z. generated the expression constructs and established the activity assays to reconstitute the minimal PKS system; G.L.C.G. and M.R. performed mass spectrometry analyses; A.B. cloned mutants, and purified and crystallized proteins; M.G. determined X-ray structures; A.B., M.S., V.R.I.K. and M.G. analysed structures; V.R.I.K. carried out molecular simulations; and M.G. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Supplementary Tables 1–13.
Rights and permissions
About this article
Cite this article
Bräuer, A., Zhou, Q., Grammbitter, G.L.C. et al. Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis. Nat. Chem. 12, 755–763 (2020). https://doi.org/10.1038/s41557-020-0491-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-020-0491-7
This article is cited by
-
C–N bond formation by a polyketide synthase
Nature Communications (2023)
-
Enzymology of assembly line synthesis by modular polyketide synthases
Nature Chemical Biology (2023)
-
Simultaneous DHA and organic selenium production by Schizochytrium sp.: a theoretical basis
Scientific Reports (2023)
-
Engineering of PKS Megaenzymes—A Promising Way to Biosynthesize High-Value Active Molecules
Topics in Catalysis (2022)
-
Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis
Scientific Reports (2021)