Abstract
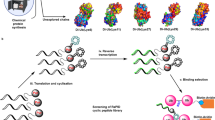
A promising approach in cancer therapy is to find ligands that directly bind ubiquitin (Ub) chains. However, finding molecules capable of tightly and specifically binding Ub chains is challenging given the range of Ub polymer lengths and linkages and their subtle structural differences. Here, we use total chemical synthesis of proteins to generate highly homogeneous Ub chains for screening against trillion-member macrocyclic peptide libraries (RaPID system). De novo cyclic peptides were found that can bind tightly and specifically to K48-linked Ub chains, confirmed by NMR studies. These cyclic peptides protected K48-linked Ub chains from deubiquitinating enzymes and prevented proteasomal degradation of Ub-tagged proteins. The cyclic peptides could enter cells, inhibit growth and induce programmed cell death, opening new opportunities for therapeutic intervention. This highly synthetic approach, with both protein target generation and cyclic peptide discovery performed in vitro, will make other elaborate post-translationally modified targets accessible for drug discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Glickman, M. H. & Ciechanover, A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 (2002).
Hersko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Pickart, C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (2001).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Gopinath, P., Ohayon, S., Nawatha, M. & Brik, A. Chemical and semisynthetic approaches to study and target deubiquitinases. Chem. Soc. Rev. 45, 4171–4198 (2016).
Ikeda, F. & Dikic, I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 536–542 (2008).
Finley, D. Recognition and processing of ubiquitin–protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009).
Reyes-Turcu, F. E. & Wilkinson, K. D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 109, 1495–1508 (2009).
Huang, X. & Dixit, V. M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016).
Pickart, C. M. & VanDemark, A. P. Opening doors into the proteasome. Nat. Struct. Biol. 7, 999–1001 (2000).
Adams, J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5, 417–421 (2004).
Goldberg, A. L. Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588 (2012).
Groll, M., Berkers, C. R., Ploegh, H. L. & Ovaa, H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 14, 451–456 (2006).
Richardson, P. G. et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med. 352, 2487–2498 (2005).
Deshaies, R. J. Proteotoxic crisis, the ubiquitin–proteasome system, and cancer therapy. BMC Biol. 12, 94 (2014).
Lee, D. H. & Goldberg, A. L. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403 (1998).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–77 (2018).
Verma, R. et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 306, 117–120 (2004).
Nakasone, M. A. et al. Structural basis for the inhibitory effects of ubistatins in the ubiquitin–proteasome pathway. Structure 25, 1839–1855 (2017).
Ye, Y. et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492, 266–270 (2012).
Castañeda, C. A. et al. Linkage-specific conformational ensembles of non-canonical polyubiquitin chains. Phys. Chem. Chem. Phys. 18, 5771–5788 (2016).
Jongkees, Sa. K., Hipolito, C. J., Rogers, J. M. & Suga, H. Model foldamers: applications and structures of stable macrocyclic peptides identified using in vitro selection. New J. Chem. 39, 3197–3207 (2015).
Zorzi, A., Deyle, K. & Heinis, C. Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol. 38, 24–29 (2017).
Goto, Y., Katoh, T. & Suga, H. Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 (2011).
Goto, Y. et al. Reprogramming the initiation event in translation for the synthesis of physiologically stable cyclic peptides. ACS Chem. Biol. 3, 120–129 (2008).
Yamagishi, Y. et al. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem. Biol. 18, 1562–1570 (2011).
Jongkees, S. A. K. et al. Rapid discovery of potent and selective glycosidase-inhibiting de novo peptides. Cell. Chem. Biol. 24, 381–390 (2017).
Mali, S. M., Singh, S. K., Eid, E. & Brik, A. Ubiquitin signaling: chemistry comes to the rescue. J. Am. Chem. Soc. 139, 4971–4986 (2017).
Orlowski, R. Z. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 6, 303 (1999).
Maki, C. G., Huibregtse, J. M. & Howley, P. M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 56, 2649–2654 (1996).
Li, B. & Dou, Q. P. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc. Natl Acad. Sci. USA 97, 3850–3855 (2000).
Lloyd, R. V. et al. P27Kip1: a multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am. J. Pathol. 154, 313–323 (1999).
Vaziri, S. A. J. et al. Inhibition of proteasome activity by bortezomib in renal cancer cells is p53 dependent and VHL independent. Anticancer Res. 29, 2961–2969 (2009).
Ajish Kumar, K. S., Haj-Yahya, M., Olschewski, D., Lashuel, H. A. & Brik, A. Highly efficient and chemoselective peptide ubiquitylation. Angew. Chem. Int. Ed. 48, 8090–8094 (2009).
Bavikar, S. N. et al. Chemical synthesis of ubiquitinated peptides with varying lengths and types of ubiquitin chains to explore the activity of deubiquitinases. Angew. Chem. Int. Ed. 51, 758–763 (2012).
Eddins, M. J., Varadan, R., Fushman, D., Pickart, C. M. & Wolberger, C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367, 204–211 (2007).
Varadan, R., Assfalg, M., Raasi, S., Pickart, C. & Fushman, D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 18, 687–698 (2005).
Mevissen, T. E. T. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013).
Renatus, M. et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 14, 1293–1302 (2006).
Stanley, M. & Virdee, S. Chemical ubiquitination for decrypting a cellular code. Biochem. J. 473, 1297–1314 (2016).
Bremm, A., Freund, S. M. V. & Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 17, 939–947 (2010).
Pagano, M. et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269, 682–685 (1995).
Devine, T. & Dai, M.-S. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr. Pharm. Des. 19, 3248–3262 (2013).
Raasi, S., Varadan, R., Fushman, D. & Pickart, C. M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12, 708–714 (2005).
Newton, K. et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008).
Pye, C. R. et al. Nonclassical size dependence of permeation defines bounds for passive adsorption of large drug molecules. J. Med. Chem. 60, 1665–1672 (2017).
Lü, S. & Wang, J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomark. Res. 1, 13 (2013).
Roscoe, B. P., Thayer, K. M., Zeldovich, K. B., Fushman, D. & Bolon, D. N. A. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol. 425, 1363–1377 (2013).
Acknowledgements
A.B. holds the Jordan and Irene Tark Academic Chair. Ha.S. is supported at the Technion by a Technion-Guangdong Fellowship. This work was supported by the Japan Agency for Medical Research and Development, Basic Science and Platform Technology Programme for Innovative Biological Medicine (JP18am0301001) to Hi.S., and by NIH grant GM065334 to D.F. J.M.R. was supported by Grants-in-aid for JSPS Fellows (P13766) and a joint ANR-JST grant (ANR-14-JITC-2014-003 and JST-SICORP). The authors thank A. Majumdar for help with triple-resonance NMR experiments. A.C. is supported by the Dr Miriam and Sheldon Adelson Medical Research Foundation (AMRF), the Israel Science Foundation (ISF), the German–Israeli Foundation for Research and Development (GIF) and a Professorship funded by the Israel Cancer Research Fund (ICRF).
Author information
Authors and Affiliations
Contributions
J.M.R. utilized the cyclic peptide discovery RaPID system, carried out SPR assays and data analysis, and co-wrote the paper. M.N. assisted in the chemical synthesis of cyclic peptides, carried out in vitro and cellular assays and co-wrote the paper. I.L. carried out the confocal microscopy assay and assisted with cellular studies. S.M.B. and B.L. synthesized isotope-labelled Ub chains, conducted the NMR studies and assisted with writing the paper. S.M.M. assisted with chemical synthesis of the ubiquitin chains. G.B.V. assisted in the synthesis of cyclic peptides. Ha.S. prepared the δ-mercaptolysine used in the ubiquitin chain synthesis. B.B. assisted with the design of the in vitro proteasomal degradation assay. D.F. designed and supervised the NMR studies, carried out data analysis and assisted with writing the manuscript and the Supplementary Information. Y.H. performed additional SPR studies against K11- and K63-linked Ub chains. A.C. assisted in the design of the confocal microscopy assay and in vitro and cellular studies. Hi.S. supervised the RaPID study and assisted in the writing of the paper. A.B. designed and supervised the entire project and the writing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20. Supplementary methods. Supplementary Table 1. Supplementary references 1–14
Rights and permissions
About this article
Cite this article
Nawatha, M., Rogers, J.M., Bonn, S.M. et al. De novo macrocyclic peptides that specifically modulate Lys48-linked ubiquitin chains. Nat. Chem. 11, 644–652 (2019). https://doi.org/10.1038/s41557-019-0278-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-019-0278-x
This article is cited by
-
DCAF13 inhibits the p53 signaling pathway by promoting p53 ubiquitination modification in lung adenocarcinoma
Journal of Experimental & Clinical Cancer Research (2024)
-
Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation
Nature Communications (2023)
-
Selective macrocyclic peptide modulators of Lys63-linked ubiquitin chains disrupt DNA damage repair
Nature Communications (2022)
-
A context-dependent and disordered ubiquitin-binding motif
Cellular and Molecular Life Sciences (2022)