Abstract
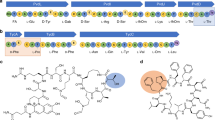
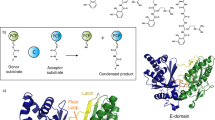
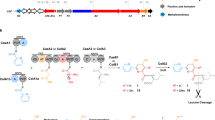
Non-ribosomal peptide synthetases (NRPSs) are giant enzyme machines that activate amino acids in an assembly line fashion. As NRPSs are not restricted to the incorporation of the 20 proteinogenic amino acids, their efficient manipulation would enable microbial production of a diverse range of peptides; however, the structural requirements for reprogramming NRPSs to facilitate the production of new peptides are not clear. Here we describe a new fusion point inside the condensation domains of NRPSs that results in the development of the exchange unit condensation domain (XUC) concept, which enables the efficient production of peptides, even containing non-natural amino acids, in yields up to 280 mg l−1. This allows the generation of more specific NRPSs, reducing the number of unwanted peptide derivatives, but also the generation of peptide libraries. The XUC might therefore be suitable for the future optimization of peptide production and the identification of bioactive peptide derivatives for pharmaceutical and other applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Clardy, J., Fischbach, M. A. & Walsh, C. T. New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550 (2006).
von Nussbaum, F., Brands, M., Hinzen, B., Weigand, S. & Häbich, D. Antibacterial natural products in medicinal chemistry—exodus or revival? Angew. Chem. Int. Ed. 45, 5072–5129 (2006).
Nathan, C. Antibiotics at the crossroads. Nature 431, 899–902 (2004).
Felnagle, E. A. et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharmaceut. 5, 191–211 (2008).
Calcott, M. J. & Ackerley, D. F. Genetic manipulation of non-ribosomal peptide synthetases to generate novel bioactive peptide products. Biotechnol. Lett. 36, 2407–2416 (2014).
Sieber, S. A. & Marahiel, M. A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005).
O’Connell, K. M. G. et al. Combating multidrug-resistant bacteria: current strategies for the discovery of novel antibacterials. Angew. Chem. Int. Ed. 52, 10706–10733 (2013).
Bush, K. Improving known classes of antibiotics: an optimistic approach for the future. Curr. Opin. Pharmacol. 12, 527–534 (2012).
Kirschning, A. & Hahn, F. Merging chemical synthesis and biosynthesis: a new chapter in the total synthesis of natural products and natural product libraries. Angew. Chem. Int. Ed. 51, 4012–4022 (2012).
Baltz, R. H. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth. Biol. 3, 748–758 (2014).
Winn, M., Fyans, J. K., Zhuo, Y. & Micklefield, J. Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 33, 317–347 (2016).
Caboche, S., Leclere, V., Pupin, M., Kucherov, G. & Jacques, P. Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. J. Bacteriol. 192, 5143–5150 (2010).
Grünewald, J. & Marahiel, M. A. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70, 121–146 (2006).
Cane, D. E., Walsh, C. T. & Khosla, C. Harnessing the biosynthetic code: combinations, permutations and mutations. Science 282, 63–68 (1998).
Mootz, H. D., Schwarzer, D. & Marahiel, M. A. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3, 490–504 (2002).
Süssmuth, R. D. & Mainz, A. Nonribosomal peptide synthesis—principles and prospects. Angew. Chem. Int. Ed. 56, 3770–3821 (2017).
Balibar, C. J., Vaillancourt, F. H. & Walsh, C. T. Generation of d amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 12, 1189–1200 (2005).
Stachelhaus, T., Schneider, A. & Marahiel, M. A. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 269, 69–72 (1995).
Calcott, M. J., Owen, J. G., Lamont, I. L. & Ackerley, D. F. Biosynthesis of novel pyoverdines by domain substitution in a nonribosomal peptide synthetase of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 80, 5723–5731 (2014).
Thirlway, J. et al. Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity. Angew. Chem. Int. Ed. 51, 7181–7184 (2012).
Kries, H. et al. Reprogramming nonribosomal peptide synthetases for ‘clickable’ amino acids. Angew. Chem. Int. Ed. 53, 10105–10108 (2014).
Nguyen, K. T. et al. Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc. Natl Acad. Sci. USA 103, 17462–17467 (2006).
Yakimov, M. M., Giuliano, L., Timmis, K. N. & Golyshin, P. N. Recombinant acylheptapeptide lichenysin: high level of production by Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol. 2, 217–224 (2000).
Beer, R. et al. Creating functional engineered variants of the single-module non-ribosomal peptide synthetase IndC by T domain exchange. Mol. BioSyst. 10, 1709–1710 (2014).
Calcott, M. J. & Ackerley, D. F. Portability of the thiolation domain in recombinant pyoverdine non-ribosomal peptide synthetases. BMC Microbiol. 15, 1–13 (2015).
Chiocchini, C., Linne, U. & Stachelhaus, T. In vivo biocombinatorial synthesis of lipopeptides by COM domain-mediated reprogramming of the surfactin biosynthetic complex. Chem. Biol. 13, 899–908 (2006).
Duerfahrt, T., Doekel, S., Sonke, T., Quaedflieg, P. J. L. M. & Marahiel, M. A. Construction of hybrid peptide synthetases for the production of alpha-l-aspartyl-l-phenylalanine, a precursor for the high-intensity sweetener aspartame. Eur. J. Biochem. 270, 4555–4563 (2003).
Brown, A. S., Calcott, M. J., Owen, J. G. & Ackerley, D. F. Structural, functional and evolutionary perspectives on effective re-engineering of non-ribosomal peptide synthetase assembly lines. Nat. Prod. Rep. 49, 104–119 (2018).
Kries, H. Biosynthetic engineering of nonribosomal peptide synthetases. J. Pept. Sci. 22, 564–570 (2016).
Bozhüyük, K. A. J. et al. De novo design and engineering of non-ribosomal peptide synthetases. Nat. Chem. 10, 275–281 (2018).
Linne, U. & Marahiel, M. A. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry 39, 10439–10447 (2000).
Bode, H. B. et al. Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chem. Eur. J. 18, 2342–2348 (2012).
Nollmann, F. I. et al. Insect-specific production of new GameXPeptides in Photorhabdus luminescens TTO1, widespread natural products in entomopathogenic bacteria. ChemBioChem 16, 205–208 (2015).
Fuchs, S. W. et al. Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal. Chem. 84, 6948–6955 (2012).
Samel, S. A., Schoenafinger, G., Knappe, T. A., Marahiel, M. A. & Essen, L.-O. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure 15, 781–792 (2007).
Keating, T. A., Marshall, C. G., Walsh, C. T. & Keating, A. E. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat. Struct. Biol. 9, 522–526 (2002).
Tanovic, A., Samel, S. A., Essen, L.-O. & Marahiel, M. A. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science 321, 659–663 (2008).
Bloudoff, K., Rodionov, D. & Schmeing, T. M. Crystal structures of the first condensation domain of CDA synthetase suggest conformational changes during the synthetic cycle of nonribosomal peptide synthetases. J. Mol. Biol. 425, 3137–3150 (2013).
Rausch, C., Hoof, I., Weber, T., Wohlleben, W. & Huson, D. H. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 7, 78 (2007).
Marahiel, M. A. A structural model for multimodular NRPS assembly lines. Nat. Prod. Rep. 33, 136–140 (2016).
Konz, D., Klens, A., Schörgendorfer, K. & Marahiel, M. A. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4, 927–937 (1997).
Cosmina, P. et al. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8, 821–831 (1993).
Krätzschmar, J., Krause, M. & Marahiel, M. A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J. Bacteriol. 171, 5422–5429 (1989).
Mootz, H. D. & Marahiel, M. A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179, 6843–6850 (1997).
Li, R., Oliver, R. A. & Townsend, C. A. Identification and characterization of the sulfazecin monobactam biosynthetic gene cluster. Cell Chem. Biol. 24, 24–34 (2017).
Meyer, S. et al. Biochemical dissection of the natural diversification of microcystin provides lessons for synthetic biology of NRPS. Cell Chem. Biol. 23, 462–471 (2016).
Schimming, O., Fleischhacker, F., Nollmann, F. I. & Bode, H. B. Yeast homologous recombination cloning leading to the novel peptides ambactin and xenolindicin. ChemBioChem 15, 1290–1294 (2014).
Roy, A. D., Grüschow, S., Cairns, N. & Goss, R. J. M. Gene expression enabling synthetic diversification of natural products: chemogenetic generation of pacidamycin analogs. J. Am. Chem. Soc. 132, 12243–12245 (2010).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click‐chemie: diverse chemische Funktionalität mit einer Handvoll guter Reaktionen. Angew. Chem. 113, 2056–2075 (2001).
Pérez, A. J., Wesche, F., Adihou, H. & Bode, H. B. Solid-phase enrichment and analysis of azide-labeled natural products: fishing downstream of biochemical pathways. Chem. Eur. J. 22, 639–645 (2016).
Pérez, A. J. & Bode, H. B. ‘Click chemistry’ for the simple determination of fatty-acid uptake and degradation: revising the role of fatty-acid transporters. ChemBioChem 16, 1588–1591 (2015).
Kronenwerth, M. et al. Characterisation of taxlllaids A-G; natural products from Xenorhabdus indica. Chem. Eur. J. 20, 17478–17487 (2014).
Phelan, V. V., Du, Y., McLean, J. A. & Bachmann, B. O. Adenylation enzyme characterization using gamma-(18)O(4)-ATP pyrophosphate exchange. Chem. Biol. 16, 473–478 (2009).
Kegler, C. et al. Rapid determination of the amino acid configuration of xenotetrapeptide. ChemBioChem 15, 826–828 (2014).
Harvey, A. L., Edrada-Ebel, R. & Quinn, R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14, 111–129 (2015).
Gietz, R. D. & Schiestl, R. H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 1–4 (2007).
Bode, H. B. et al. Structure elucidation and activity of kolossin A, the d-/l-pentadecapeptide product of a giant nonribosomal peptide synthetase. Angew. Chem. Int. Ed. 54, 10352–10355 (2015).
Fuchs, S. W., Proschak, A., Jaskolla, T. W., Karas, M. & Bode, H. B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 9, 3130–3132 (2011).
Horsman, M. E., Hari, T. P. A. & Boddy, C. N. Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat. Prod. Rep. 33, 183–202 (2016).
Acknowledgements
The authors thank M. Lindner and C. Zizka for help with the construction of selected plasmids and C. Kegler for helpful discussions. This work was funded in part by the LOEWE programme of the state of Hesse as part of the MegaSyn and TBG research clusters. H.B.B. acknowledges the Deutsche Forschungsgemeinschaft for funding of the Impact II qTof mass spectrometer (INST 161/810-1).
Author information
Authors and Affiliations
Contributions
K.A.J.B. and H.B.B. designed the experiments. K.A.J.B., A.L., A.T., J.K., S.N. and F.F. performed all molecular biology and biochemical experiments. F.W. synthesized all peptide standards that were used for the high-performance liquid chromatography–mass spectrometry-based quantification performed by A.L. and A.T. J.K., Y.-N.S. and P.G. isolated selected peptides and Y.-N.S. performed their NMR analysis. All authors analysed the results and K.A.J.B., A.L., A.T., J.K. and H.B.B. wrote the manuscript. All authors saw and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Information
Supplementary Information includes methods, Supplementary Tables 1–7 (containing mass spectrometry data of the produced peptides, strains, plasmids and oligonucleotides used in this study) and Supplementary Figs. 1–42 (including NMR spectra of new peptides).
Rights and permissions
About this article
Cite this article
Bozhüyük, K.A.J., Linck, A., Tietze, A. et al. Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 11, 653–661 (2019). https://doi.org/10.1038/s41557-019-0276-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-019-0276-z
This article is cited by
-
High-throughput reprogramming of an NRPS condensation domain
Nature Chemical Biology (2024)
-
Metagenomic domain substitution for the high-throughput modification of nonribosomal peptides
Nature Chemical Biology (2024)
-
Genetically programmed cell-based synthesis of non-natural peptide and depsipeptide macrocycles
Nature Chemistry (2023)
-
Resurrecting ancestral antibiotics: unveiling the origins of modern lipid II targeting glycopeptides
Nature Communications (2023)
-
Bifurcation drives the evolution of assembly-line biosynthesis
Nature Communications (2022)