Abstract
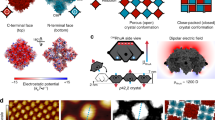
De novo design and construction of stimuli-responsive protein assemblies that predictably switch between discrete conformational states remains an essential but highly challenging goal in biomolecular design. We previously reported synthetic, two-dimensional protein lattices self-assembled via disulfide bonding interactions, which endows them with a unique capacity to undergo coherent conformational changes without losing crystalline order. Here, we carried out all-atom molecular dynamics simulations to map the free-energy landscape of these lattices, validated this landscape through extensive structural characterization by electron microscopy and established that it is predominantly governed by solvent reorganization entropy. Subsequent redesign of the protein surface with conditionally repulsive electrostatic interactions enabled us to predictably perturb the free-energy landscape and obtain a new protein lattice whose conformational dynamics can be chemically and mechanically toggled between three different states with varying porosities and molecular densities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Marsh, J. A. & Teichmann, S. A. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 84, 551–575 (2015).
Salgado, E. N., Radford, R. J. & Tezcan, F. A. Metal-directed protein self-assembly. Acc. Chem. Res. 43, 661–672 (2010).
Song, W. J. & Tezcan, F. A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014).
Yeates, T. O. Geometric principles for designing highly symmetric self-assembling protein nanomaterials. Annu. Rev. Biophys. 46, 23–42 (2017).
Bale, J. B. et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016).
Koga, N. et al. Principles for designing ideal protein structures. Nature 491, 222–227 (2012).
Norn, C. H. & André, I. Computational design of protein self-assembly. Curr. Opin. Struct. Biol. 39, 39–45 (2016).
Ringler, P. & Schulz, G. E. Self-assembly of proteins into designed networks. Science 302, 106–109 (2003).
Sinclair, J. C., Davies, K. M., Venien-Bryan, C. & Noble, M. E. M. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat. Nanotechnol. 6, 558–562 (2011).
Levy, Y. & Onuchic, J. N. Water mediation in protein folding and molecular recognition. Annu. Rev. Biophys. Biomol. Struct. 35, 389–415 (2006).
Young, T., Abel, R., Kim, B., Berne, B. J. & Friesner, R. A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein–ligand binding. Proc. Natl Acad. Sci. USA 104, 808–813 (2007).
Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 (2002).
Davey, J. A., Damry, A. M., Goto, N. K. & Chica, R. A. Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 13, 1280–1285 (2017).
Joh, N. H. et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014).
Cerasoli, E., Sharpe, B. K. & Woolfson, D. N. ZiCo: a peptide designed to switch folded state upon binding zinc. J. Am. Chem. Soc. 127, 15008–15009 (2005).
Ambroggio, X. I. & Kuhlman, B. Computational design of a single amino acid sequence that can switch between two distinct protein folds. J. Am. Chem. Soc. 128, 1154–1161 (2006).
Brodin, J. D. et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem. 4, 375–382 (2012).
Sontz, P. A., Bailey, J. B., Ahn, S. & Tezcan, F. A. A metal organic framework with spherical protein nodes: rational chemical design of 3D protein crystals. J. Am. Chem. Soc. 137, 11598–11601 (2015).
Bailey, J. B., Subramanian, R. H., Churchfield, L. A. & Tezcan, F. A. Metal-directed design of supramolecular protein assemblies. Methods Enzymol. 580, 223–250 (2016).
Suzuki, Y. et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016).
Grima, J. N. & Evans, K. E. Auxetic behavior from rotating squares. J. Mater. Sci. Lett. 19, 1563–1565 (2000).
Yang, W., Li, Z.-M., Shi, W., Xie, B.-H. & Yang, M.-B. Review on auxetic materials. J. Mater. Sci. 39, 3269–3279 (2004).
Evans, K. E. & Alderson, A. Auxetic materials: functional materials and structures from lateral thinking! Adv. Mater. 12, 617–628 (2000).
Zhu, F. & Hummer, G. Convergence and error estimation in free energy calculations using the weighted histogram analysis method. J. Comput. Chem. 33, 453–465 (2012).
Kumar, S., Rosenberg, J. M., Bouzida, D., Swendsen, R. H. & Kollman, P. A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992).
De Yoreo, J. J. & Vekilov, P. G. Principles of crystal nucleation and growth. Rev. Mineral. Geochem. 54, 57–93 (2003).
Derewenda, Z. S. & Vekilov, P. G. Entropy and surface engineering in protein crystallization. Acta Crystallogr. D 62, 116–124 (2006).
Nguyen, C. N., Young, T. K. & Gilson, M. K. Grid inhomogeneous solvation theory: hydration structure and thermodynamics of the miniature receptor cucurbit[7]uril. J. Chem. Phys. 137, 044101 (2012).
Huggins, D. J. Comparing distance metrics for rotation using the k-nearest neighbors algorithm for entropy estimation. J. Comput. Chem. 35, 377–385 (2014).
Giovambattista, N., Rossky, P. J. & Debenedetti, P. G. Effect of temperature on the structure and phase behavior of water confined by hydrophobic, hydrophilic, and heterogeneous surfaces. J. Phys. Chem. B 113, 13723–13734 (2009).
Tarek, M. & Tobias, D. J. The dynamics of protein hydration water: a quantitative comparison of molecular dynamics simulations and neutron-scattering experiments. Biophys. J. 79, 3244–3257 (2000).
Ebbinghaus, S. et al. An extended dynamical hydration shell around proteins. Proc. Natl Acad. Sci. USA 104, 20749–20752 (2007).
Heyden, M. et al. Dissecting the THz spectrum of liquid water from first principles via correlations in time and space. Proc. Natl Acad. Sci. USA 107, 12068–12073 (2010).
Dunitz, J. D. The entropic cost of bound water in crystals and biomolecules. Science 264, 670 (1994).
Yuan, Y., Tam, M. F., Simplaceanu, V. & Ho, C. New look at hemoglobin allostery. Chem. Rev. 115, 1702–1724 (2015).
Joshi, R. K. et al. Precise and ultrafast molecular sieving through graphene oxide membranes. Science 343, 752–754 (2014).
Tahara, K., Nakatani, K., Iritani, K., De Feyter, S. & Tobe, Y. Periodic functionalization of surface-confined pores in a two-dimensional porous network using a tailored molecular building block. ACS Nano 10, 2113–2120 (2016).
Huber, C. et al. Heterotetramers formed by an S-layer–streptavidin fusion protein and core-streptavidin as a nanoarrayed template for biochip development. Small 2, 142–150 (2006).
Li, Y. et al. MoS2 nanoparticles grown on graphene: an advanced catalyst for the hydrogen evolution reaction. J. Am. Chem. Soc. 133, 7296–7299 (2011).
Mader, C., Küpcü, S., Sleytr, U. B. & Sára, M. S-layer-coated liposomes as a versatile system for entrapping and binding target molecules. Biochim. Biophys. Acta Biomembr. 1463, 142–150 (2000).
Butler, S. Z. et al. Progress, challenges, and opportunities in two-dimensional materials beyond graphene. ACS Nano 7, 2898–2926 (2013).
Acknowledgements
We thank M. Gilson, T. Kurtzman and S. Ramsey for helpful discussions regarding GIST, R. Subramanian for assistance with the generation of projection maps from computational models, and T. Baker for use of the electron microscopy facilities. This work was primarily supported by the US Department of Energy (Division of Materials Sciences, Office of Basic Energy Sciences; Award DE-SC0003844 to F.A.T.). F.P. was supported by the National Science Foundation through grant CHE-1453204 (computation). R.A. was supported in part by the University of California, San Diego NIH Molecular Biophysics Training Grant (T32-GM08326). All computer simulations were performed on the Extreme Science and Engineering Discovery Environment, which is supported by the National Science Foundation through grant ACI-1053575.
Author information
Authors and Affiliations
Contributions
R.A. co-initiated the project, designed and performed all of the experiments, simulations and data analysis, and co-wrote the paper. Y.S. performed the TEM data collection and analysis. F.P. guided the simulation design and computational data analysis, and co-wrote the paper. F.A.T. initiated the project, guided the experiment design and data analysis, and co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Methods, Supplementary Figures 1–11, Supplementary Table 1 and Supplementary References
Rights and permissions
About this article
Cite this article
Alberstein, R., Suzuki, Y., Paesani, F. et al. Engineering the entropy-driven free-energy landscape of a dynamic nanoporous protein assembly. Nature Chem 10, 732–739 (2018). https://doi.org/10.1038/s41557-018-0053-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0053-4
This article is cited by
-
Design of supramolecular nanosheets for drug delivery applications
Polymer Journal (2023)
-
Structural resolution of switchable states of a de novo peptide assembly
Nature Communications (2021)
-
Design of metal-mediated protein assemblies via hydroxamic acid functionalities
Nature Protocols (2021)
-
Asymmetrizing an icosahedral virus capsid by hierarchical assembly of subunits with designed asymmetry
Nature Communications (2021)
-
Design of biologically active binary protein 2D materials
Nature (2021)