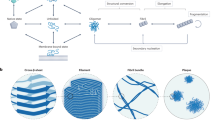
Abstract
Mapping free-energy landscapes has proved to be a powerful tool for studying reaction mechanisms. Many complex biomolecular assembly processes, however, have remained challenging to access using this approach, including the aggregation of peptides and proteins into amyloid fibrils implicated in a range of disorders. Here, we generalize the strategy used to probe free-energy landscapes in protein folding to determine the activation energies and entropies that characterize each of the molecular steps in the aggregation of the amyloid-β peptide (Aβ42), which is associated with Alzheimer’s disease. Our results reveal that interactions between monomeric Aβ42 and amyloid fibrils during fibril-dependent secondary nucleation fundamentally reverse the thermodynamic signature of this process relative to primary nucleation, even though both processes generate aggregates from soluble peptides. By mapping the energetic and entropic contributions along the reaction trajectories, we show that the catalytic efficiency of Aβ42 fibril surfaces results from the enthalpic stabilization of adsorbing peptides in conformations amenable to nucleation, resulting in a dramatic lowering of the activation energy for nucleation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Sipe, J. D. et al. Amyloid fibril protein nomenclature: 2012 recommendations from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid 19, 167–170 (2012).
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000).
Knowles, T. P. J. et al. An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009).
Lee, J., Culyba, E. K., Powers, E. T. & Kelly, J. W. Amyloid-beta forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 7, 602–609 (2011).
Cohen, S. I. A. et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA 110, 9758–9763 (2013).
Meisl, G. et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl Acad. Sci. USA 111, 9384–9389 (2014).
Cohen, S. I. A. et al. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat. Struct. Mol. Biol. 22, 207–213 (2015).
Hellstrand, E., Boland, B., Walsh, D. M. & Linse, S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem. Neurosci. 1, 13–18 (2010).
Cohen, S. I. A. et al. Nucleated polymerization with secondary pathways. I. Time evolution of the principal moments. J. Chem. Phys. 135, 065105 (2011).
Kar, K., Jayaraman, M., Sahoo, B., Kodali, R. & Wetzel, R. Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat. Struct. Mol. Biol. 18, 328–336 (2011).
Oosawa, F. & Asakura, S. Thermodynamics of the Polymerization of Protein (Academic Press, New York, NY, 1975).
Jarrett, J. T. & Lansbury, P. T. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in alzheimer’s disease and scrapie? Cell 73, 1055–1058 (1993).
Collins, S. R., Douglass, A., Vale, R. D. & Weissman, J. S. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2, e321 (2004).
Jeong, J. S., Ansaloni, A., Mezzenga, R., Lashuel, H. A. & Dietler, G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J. Mol. Biol. 425, 1765–1781 (2013).
Ruschak, A. M. & Miranker, A. D. Fiber-dependent amyloid formation as catalysis of an existing reaction pathway. Proc. Natl Acad. Sci. USA 104, 12341–12346 (2007).
Buell, A. K. et al. Solution conditions determine the relative importance of nucleation and growth processes in alpha-synuclein aggregation. Proc. Natl Acad. Sci. USA 111, 7671–7676 (2014).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Haass, C. & Selkoe, D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell. Biol. 8, 101–112 (2007).
Walsh, D. M. et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002).
Bucciantini, M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002).
Knowles, T. P. J. et al. Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl Acad. Sci. USA 108, 14746–14751 (2011).
Cohen, S. I. A. et al. Spatial propagation of protein polymerization. Phys. Rev. Lett. 112, 098101 (2014).
Jucker, M. & Walker, L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Ferrone, F. Analysis of protein aggregation kinetics. Methods Enzymol. 309, 256–274 (1999).
Cohen, S. I. A., Vendruscolo, M., Dobson, C. M. & Knowles, T. P. J. Nucleated polymerization with secondary pathways. II. Determination of self-consistent solutions to growth processes described by non-linear master equations. J. Chem. Phys. 135, 065106 (2011).
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Schuler, B., Lipman, E. A. & Eaton, W. A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 419, 743–747 (2002).
Kramers, H. A. Brownian motion in a field of forceand the diffusion model of chemical reactions. Physica 7, 284 (1940).
Zwanzig, R. Two-state models of protein folding kinetics. Proc. Natl Acad. Sci. USA 94, 148–150 (1997).
Buell, A. K. et al. Frequency factors in a landscape model of filamentous protein aggregation. Phys. Rev. Lett. 104, 228101 (2010).
Buell, A. K. et al. Detailed analysis of the energy barriers for amyloid fibril growth. Angew. Chem. Int. Ed. 51, 5247–5251 (2012).
Zwanzig, R. Diffusion in a rough potential. Proc. Natl Acad. Sci. USA 85, 2029–2030 (1988).
Knowles, T. P. J. et al. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc. Natl Acad. Sci. USA 104, 10016–10021 (2007).
Kashchiev, D. & Auer, S. Nucleation of amyloid fibrils. J. Chem. Phys. 132, 215101 (2010).
Mozurkewich, M. & Benson, S. W. Negative activation energies and curved arrhenius plots. 1. theory of reactions over potential wells. J. Phys. Chem. 88, 6429–6435 (1984).
Oliveberg, M., Tan, Y. J. & Fersht, A. R. Negative activation enthalpies in the kinetics of protein folding. Proc. Natl Acad. Sci. USA 92, 8926–8929 (1995).
Saric, A. et al. Physical determinants of the self-replication of protein fibrils. Nat. Phys. 12, 874–880 (2016).
Roduner, E. Understanding catalysis. Chem. Soc. Rev. 43, 8226–8239 (2014).
Medford, A. et al. From the Sabatier principle to a predictive theory of transition-metal heterogeneous catalysis. J. Cat. 328, 36–42 (2015).
Anwar, J., Khan, S. & Lindfors, L. Secondary crystal nucleation: nuclei breeding factory uncovered. Angew. Chem. Int. Ed. 54, 14681–14684 (2015).
Cukalevski, R. et al. The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem. Sci. 6, 4215–4233 (2015).
Walsh, D. M. et al. A facile method for expression and purification of the Alzheimer’s disease-associated amyloid beta-peptide. FEBS J. 276, 1266–1281 (2009).
Lührs, T. et al. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc. Natl Acad. Sci. USA 102, 17342–17347 (2005).
Crescenzi, O. et al. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. similarity with a virus fusion domain. Eur. J. Biochem. 269, 5642–5648 (2002).
Young, L. J., Kaminski Schierle, G. S. & Kaminski, C. F. Imaging Aβ(1-42) fibril elongation reveals strongly polarised growth and growth incompetent states. Phys. Chem. Chem. Phys. 19, 27987–27996 (2017).
Acknowledgements
We thank B. Jönsson and I. André for helpful discussions. We acknowledge financial support from the Schiff Foundation (S.I.A.C.), St John’s College, Cambridge (S.I.A.C.), the Royal Physiographic Society (R.C.), the Research School FLÄK of Lund University (S.L., R.C.), the Swedish Research Council (S.L.) and its Linneaus Centre Organizing Molecular Matter (S.L.), the Crafoord Foundation (S.L.), Alzheimerfonden (S.L.), the European Research Council (S.L.), NanoLund (S.L.), Knut and Alice Wallenberg Foundation (S.L.), Peterhouse, Cambridge (T.C.T.M.), the Swiss National Science Foundation (T.C.T.M.), Magdalene College, Cambridge (A.K.B.), the Leverhulme Trust (A.K.B.), the Royal Society (A.Š.), the Academy of Medical Sciences (A.Š.), the Wellcome Trust (C.M.D., T.P.J.K., A.Š.), and the Centre for Misfolding Diseases (C.M.D., T.P.J.K, M.V.). A.K.B. thanks the Alzheimer Forschung Initiative (AFI).
Author information
Authors and Affiliations
Contributions
S.I.A.C., R.C., T.P.J.K. and S.L. designed the study. R.C., M.T. and S.L. performed the experiments. S.I.A.C., A.K.B., T.C.T.M., A.Š. and T.P.J.K. analysed the data. All authors discussed the results and contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary Methods and Supplementary Figs. 1–6
Rights and permissions
About this article
Cite this article
Cohen, S.I.A., Cukalevski, R., Michaels, T.C.T. et al. Distinct thermodynamic signatures of oligomer generation in the aggregation of the amyloid-β peptide. Nature Chem 10, 523–531 (2018). https://doi.org/10.1038/s41557-018-0023-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-018-0023-x
This article is cited by
-
Amyloidogenic 60–71 deletion/ValThr insertion mutation of apolipoprotein A-I generates a new aggregation-prone segment that promotes nucleation through entropic effects
Scientific Reports (2023)
-
Mechanisms of enhanced aggregation and fibril formation of Parkinson’s disease-related variants of α-synuclein
Scientific Reports (2022)
-
Direct observation of heterogeneous formation of amyloid spherulites in real-time by super-resolution microscopy
Communications Biology (2022)
-
Electrical characteristics of amyloid beta peptides in vertical junctions
NPG Asia Materials (2021)
-
Gallic acid oxidation products alter the formation pathway of insulin amyloid fibrils
Scientific Reports (2020)