Abstract
The pancreatic islets are composed of discrete hormone-producing cells that orchestrate systemic glucose homeostasis. Here we identify subsets of beta cells using a single-cell transcriptomic approach. One subset of beta cells marked by high CD63 expression is enriched for the expression of mitochondrial metabolism genes and exhibits higher mitochondrial respiration compared with CD63lo beta cells. Human and murine pseudo-islets derived from CD63hi beta cells demonstrate enhanced glucose-stimulated insulin secretion compared with pseudo-islets from CD63lo beta cells. We show that CD63hi beta cells are diminished in mouse models of and in humans with type 2 diabetes. Finally, transplantation of pseudo-islets generated from CD63hi but not CD63lo beta cells into diabetic mice restores glucose homeostasis. These findings suggest that loss of a specific subset of beta cells may lead to diabetes. Strategies to reconstitute or maintain CD63hi beta cells may represent a potential anti-diabetic therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
scRNA-seq and bulk RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) database under accession number GSE203151 and are publicly available. Bulk RNA-seq data have been deposited at the GEO database under accession number GSE205023 and are publicly available. The previously published human scRNA-seq data that were re-analysed are obtained from GSE84133, GSE86469, GSE85241, GSE81608 and E-MTAB-5061. Meta-analysis is available from the Huising Lab Website (https://www.huisinglab.com/). Data from Extended Data Fig. 5a were obtained from GSE128565. Data from Extended Data Fig. 5b were obtained from GSE88778. Data from Extended Data Fig. 5c were obtained from GSE87375. Data from Extended Data Fig. 5d were obtained from GSE88779. Data from Extended Data Fig. 9l were obtained from European Nucleotide Archive (ENA), accession number PRJEB30761 (https://www.ebi.ac.uk/ena/data/view/PRJEB30761). Source data are provided with this paper. Source data for all graphs have been provided in source data and uploaded to the Figshare https://doi.org/10.6084/m9.figshare.21435972 public link.
Code availability
All bioinformatics analyses code has been deposited at GitHub and is publicly available as of the date of publication (https://github.com/abcwcm/RubioNavarro_Pancreas).
References
Festa, A., Williams, K., D’Agostino, R. Jr., Wagenknecht, L. E. & Haffner, S. M. The natural course of beta-cell function in nondiabetic and diabetic individuals: the Insulin Resistance Atherosclerosis Study. Diabetes 55, 1114–1120 (2006).
Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 6, 361–369 (2018).
Gomez-Banoy, N. et al. Adipsin preserves beta cells in diabetic mice and associates with protection from type 2 diabetes in humans. Nat. Med. 25, 1739–1747 (2019).
Tabak, A. G. et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373, 2215–2221 (2009).
Wagner, R. et al. Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat. Med. 27, 49–57 (2021).
Haythorne, E. et al. Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic beta-cells. Nat. Commun. 10, 2474 (2019).
Sharma, R. B., Landa-Galvan, H. V. & Alonso, L. C. Living dangerously: protective and harmful ER stress responses in pancreatic beta-cells. Diabetes 70, 2431–2443 (2021).
Accili, D. et al. When beta-cells fail: lessons from dedifferentiation. Diabetes Obes. Metab. 18, 117–122 (2016).
Hunter, C. S. & Stein, R. W. Evidence for loss in identity, de-differentiation, and trans-differentiation of islet beta-cells in type 2 diabetes. Front. Genet. 8, 35 (2017).
Knudsen, J. G. & Rorsman, P. Beta cell dysfunction in type 2 diabetes: drained of energy? Cell Metab. 29, 1–2 (2019).
Gutierrez, G. D., Gromada, J. & Sussel, L. Heterogeneity of the pancreatic beta cell. Front. Genet. 8, 22 (2017).
Pipeleers, D. G. Heterogeneity in pancreatic beta-cell population. Diabetes 41, 777–781 (1992).
Muller, A. et al. 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse beta cells. J. Cell Biol. 220, e202010039 (2021).
Katsuta, H. et al. Subpopulations of GFP-marked mouse pancreatic beta-cells differ in size, granularity, and insulin secretion. Endocrinology 153, 5180–5187 (2012).
Salomon, D. & Meda, P. Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Exp. Cell. Res. 162, 507–520 (1986).
Schuit, F. C., In’t Veld, P. A. & Pipeleers, D. G. Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proc. Natl Acad. Sci. USA 85, 3865–3869 (1988).
Jetton, T. L. & Magnuson, M. A. Heterogeneous expression of glucokinase among pancreatic beta cells. Proc. Natl Acad. Sci. USA 89, 2619–2623 (1992).
Kiekens, R. et al. Differences in glucose recognition by individual rat pancreatic B cells are associated with intercellular differences in glucose-induced biosynthetic activity. J. Clin. Invest. 89, 117–125 (1992).
Giordano, E., Bosco, D., Cirulli, V. & Meda, P. Repeated glucose stimulation reveals distinct and lasting secretion patterns of individual rat pancreatic B cells. J. Clin. Invest. 87, 2178–2185 (1991).
Van Schravendijk, C. F., Kiekens, R. & Pipeleers, D. G. Pancreatic beta cell heterogeneity in glucose-induced insulin secretion. J. Biol. Chem. 267, 21344–21348 (1992).
Heimberg, H. et al. Heterogeneity in glucose sensitivity among pancreatic beta-cells is correlated to differences in glucose phosphorylation rather than glucose transport. EMBO J. 12, 2873–2879 (1993).
Soria, B., Chanson, M., Giordano, E., Bosco, D. & Meda, P. Ion channels of glucose-responsive and -unresponsive beta-cells. Diabetes 40, 1069–1078 (1991).
Holz, G. G. T., Kuhtreiber, W. M. & Habener, J. F. Pancreatic beta-cells are rendered glucose-competent by the insulinotropic hormone glucagon-like peptide-1(7-37). Nature 361, 362–365 (1993).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360 e344 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394 e383 (2016).
Segerstolpe, A. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Xin, Y. et al. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc. Natl Acad. Sci. USA 113, 3293–3298 (2016).
Lawlor, N. et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 27, 208–222 (2017).
Xin, Y. et al. Pseudotime ordering of single human beta-cells reveals states of insulin production and unfolded protein response. Diabetes 67, 1783–1794 (2018).
Fang, Z. et al. Single-cell heterogeneity analysis and CRISPR screen identify key beta-cell-specific disease genes. Cell Rep. 26, 3132–3144 e3137 (2019).
Xin, Y. et al. Single-cell RNAseq reveals that pancreatic beta-cells from very old male mice have a young gene signature. Endocrinology 157, 3431–3438 (2016).
Dominguez-Gutierrez, G., Xin, Y. & Gromada, J. Heterogeneity of human pancreatic beta-cells. Mol. Metab. 27S, S7–S14 (2019).
Mawla, A. M. & Huising, M. O. Navigating the depths and avoiding the shallows of pancreatic islet cell transcriptomes. Diabetes 68, 1380–1393 (2019).
Camunas-Soler, J. et al. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab. 31, 1017–1031 e1014 (2020).
Bar, Y. & Efrat, S. The NOTCH pathway in beta-cell growth and differentiation. Vitam. Horm. 95, 391–405 (2014).
Bartolome, A., Zhu, C., Sussel, L. & Pajvani, U. B. Notch signaling dynamically regulates adult beta cell proliferation and maturity. J. Clin. Invest. 129, 268–280 (2019).
Kelly, P. et al. Rap1 promotes multiple pancreatic islet cell functions and signals through mammalian target of rapamycin complex 1 to enhance proliferation. J. Biol. Chem. 285, 15777–15785 (2010).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Kang, H. S. et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol. Cell. Biol. 29, 6366–6379 (2009).
Osipovich, A. B. et al. Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development 141, 2939–2949 (2014).
Newsholme, P., Keane, K. N., Carlessi, R. & Cruzat, V. Oxidative stress pathways in pancreatic beta-cells and insulin-sensitive cells and tissues: importance to cell metabolism, function, and dysfunction. Am. J. Physiol. Cell Physiol. 317, C420–C433 (2019).
Lee, J. H. et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes 58, 344–351 (2009).
Singh, C. K. et al. The role of sirtuins in antioxidant and redox signaling. Antioxid. Redox Signal 28, 643–661 (2018).
Baumel-Alterzon, S., Katz, L. S., Brill, G., Garcia-Ocana, A. & Scott, D. K. Nrf2: the master and captain of beta cell fate. Trends Endocrinol. Metab. 32, 7–19 (2021).
Gupta, R. K. et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J. Clin. Invest. 115, 1006–1015 (2005).
Miura, A. et al. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J. Biol. Chem. 281, 5246–5257 (2006).
Flannick, J., Johansson, S. & Njolstad, P. R. Common and rare forms of diabetes mellitus: towards a continuum of diabetes subtypes. Nat. Rev. Endocrinol. 12, 394–406 (2016).
Lee, A. H., Heidtman, K., Hotamisligil, G. S. & Glimcher, L. H. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc. Natl Acad. Sci. USA 108, 8885–8890 (2011).
Juliana, C. A. et al. A PDX1-ATF transcriptional complex governs beta cell survival during stress. Mol. Metab. 17, 39–48 (2018).
Salinno, C. et al. CD81 marks immature and dedifferentiated pancreatic beta-cells. Mol. Metab. 49, 101188 (2021).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Sachs, S. et al. Targeted pharmacological therapy restores beta-cell function for diabetes remission. Nat. Metab. 2, 192–209 (2020).
van der Meulen, T. et al. Virgin beta cells persist throughout life at a neogenic niche within pancreatic islets. Cell Metab. 25, 911–926 e916 (2017).
Qiu, W. L. et al. Deciphering pancreatic islet beta cell and alpha cell maturation pathways and characteristic features at the single-cell level. Cell Metab. 25, 1194–1205 e1194 (2017).
Farack, L. et al. Transcriptional heterogeneity of beta cells in the intact pancreas. Dev. Cell 48, 115–125 e114 (2019).
Dusaulcy, R. et al. High-fat diet impacts more changes in beta-cell compared to alpha-cell transcriptome. PLoS ONE 14, e0213299 (2019).
Backe, M. B., Moen, I. W., Ellervik, C., Hansen, J. B. & Mandrup-Poulsen, T. Iron regulation of pancreatic beta-cell functions and oxidative stress. Annu Rev. Nutr. 36, 241–273 (2016).
Talchai, C., Xuan, S., Lin, H. V., Sussel, L. & Accili, D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234 (2012).
Kim-Muller, J. Y. et al. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab. 20, 593–602 (2014).
Kim-Muller, J. Y. et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nat. Commun. 7, 12631 (2016).
Pasquier, A. et al. Lysosomal degradation of newly formed insulin granules contributes to beta cell failure in diabetes. Nat. Commun. 10, 3312 (2019).
Dorrell, C. et al. Human islets contain four distinct subtypes of beta cells. Nat. Commun. 7, 11756 (2016).
Ellenbroek, J. H. et al. Topologically heterogeneous beta cell adaptation in response to high-fat diet in mice. PLoS ONE 8, e56922 (2013).
Tersey, S. A. et al. Episodic beta-cell death and dedifferentiation during diet-induced obesity and dysglycemia in male mice. FASEB J. https://doi.org/10.1096/fj.201800150rr (2018).
Benninger, R. K. P. & Kravets, V. The physiological role of β-cell heterogeneity in pancreatic islet function. Nat. Rev. Endocrinol. 18, 9–22 (2022).
Bader, E. et al. Identification of proliferative and mature beta-cells in the islets of Langerhans. Nature 535, 430–434 (2016).
Karaca, M. et al. Exploring functional beta-cell heterogeneity in vivo using PSA-NCAM as a specific marker. PLoS ONE 4, e5555 (2009).
Wang, Y. J. et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038 (2016).
Li, J. et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 17, 178–187 (2016).
Wang, Y. J. & Kaestner, K. H. Single-cell RNA-seq of the pancreatic islets—a promise not yet fulfilled? Cell Metab. 29, 539–544 (2019).
Huotari, M. A. et al. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology 143, 4437–4446 (2002).
Doyle, M. E. & Egan, J. M. Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol. Ther. 113, 546–593 (2007).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Waltman, L. & van Eck, N. J. A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Smelt, M. J., Faas, M. M., de Haan, B. J. & de Vos, P. Pancreatic beta-cell purification by altering FAD and NAD(P)H metabolism. Exp. Diabetes Res 2008, 165360 (2008).
Acknowledgements
This work was supported by the following grants: NIH R01 DK121140 (J.C.L.), R01 DK121844 (J.C.L.), P30 DK063608 pilot feasibility grant (J.C.L.), R01 DK056626 (D.E.C.), R37 DK048873 (D.E.C.), R01 DK103046 (D.E.C.) and Human Pancreas Analysis Program for Type 1 Diabetes 2U01DK112217-02A1 (A.N.). Human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program (IIDP) at City of Hope, NIH Grant #U24DK098085 and the JDRF-funded IIDP Islet Award Initiative (J.C.L.). A.R.-N. was supported by a Maria Zambrano Research Fellowship (Spanish Ministry of Universities and European Next Generation Funds). N.G.-B. (118PMF032) and A.G. (9-22-PDFPM-01) were supported by American Diabetes Association postdoctoral fellowships. L.S. was supported by an American Heart Association (AHA) postdoctoral fellowship (908952). N.I. was supported by an AHA Postdoctoral Fellowship (19POST34380692) and an American Liver Foundation Irwin M. Arias, MD Postdoctoral Research Fellowship Award American. L.M. was supported by a Chinese Scholarship Council award. The views expressed in this manuscript are those of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
A.R.-N. and J.C.L. designed the study and wrote the manuscript with input from all authors. A.R.-N., N.G.-B., L.S., N.M.-O., M.R., B.Y., A.L., E.A.H. and E.C. performed and analysed the animal experiments. A.R.-N., N.G.-B., L.M., N.I., A.G. and A.C.N.C. developed and analysed the in vitro experiments. F.D., P.Z. and D.B., analysed scRNA-seq and bulk RNA-seq experiments. A.M.M. and M.O.H. analysed the human scRNA-seq data. C.L. and A.N. provided advice and human pancreas samples. D.E.C., S.C., J.C. and G.S.P. provided scientific input. J.C.L. conceived and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Raghavendra Mirmira, Michele Solimena and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characteristics of diet-induced obesity model used for scRNA-seq.
a-b, Percentages of fat mass (a), lean mass (b) in male mice fed with regular diet (RD, N = 7) or high fat diet (HFD, N = 8) for 6 weeks after weaning. c-e, Body weights (c), 6 hours fasting blood glucose (d), 6 hours fasting insulin (e) levels in male mice fed with RD (N = 11) or HFD (N = 12) for 6 weeks after weaning. f-g, Glucose tolerance test (GTT) was performed on RD (N = 7) and HFD (N = 8) fed male mice for 6 weeks with measurement of blood glucose concentrations (f). Area under the curve (AUC) of blood glucose levels (g). h-k, Insulin tolerance test (ITT) was performed on RD (N = 7) and HFD (N = 8) fed male mice for 7 weeks (h). AUC of blood glucose levels in h (i). ITT plotted as a % of starting glucose value (j). AUC of values plotted in j after t0 (k). l, Homeostasis model assessment for insulin resistance (HOMA-IR) index of male mice fed with RD (N = 11) or HFD (N = 12) for 6 weeks after weaning (insulin [mU/L]* glucose [mg/dL])/405. m-n, Quantification of β cell area (m) and mass (n) in RD (N = 4) and HFD (N = 5) fed male mice. o-r, Body weights (o), 6 hours fasting blood glucose (p), 6 hours fasting insulin levels (q), HOMA-IR (r) of female mice fed with regular diet (RD, N = 4) or high fat diet (HFD, N = 5) for 6 weeks after weaning. s-t, Quantification of β cell area (s) and mass (t) in RD (N = 4) and HFD (N = 5) fed female mice. u, Distribution of pancreatic islet cell clusters from RD (left chart) and HFD (right chart) fed male mice. v, UMAPs representing expression of known marker genes across the cell clusters Ins1 (beta cells), Gcg (alpha cells), Sst (delta cells), Ftl1 (endothelial cells), Ppt (gamma cells), and Cd74 (immune cells) in male mice fed with RD (left panel) and HFD (right panel). All data are presented as mean ± S.E.M. and Welch’s unpaired two-tailed t test is used for comparison. Source numerical data are available in source data.
Extended Data Fig. 2 Molecular analysis of β cell clusters.
a, Top 4 enriched biological processes for the indicated β cell clusters (p-value was calculated using two-tailed Benjamini-Hochberg). b, Heatmap showing select genes previously identified to be heterogeneously expressed in β cells in each of the indicated β cell clusters. c, Top 5 differentially expressed genes (DEGs) for the indicated β cell clusters. d, Pathway enrichment analysis of DEGs in Cluster 0, Cluster 2, and Cluster 3 β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). e, Top 5 transcription factors predicted in Cluster 0, Cluster 2, and Cluster 3 β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). f, Trajectory of the different β cell clusters predicted by Slingshot analysis. Source numerical data are available in source data.
Extended Data Fig. 3 CD63 expression in pancreatic islet cells.
a, Representative immunofluorescence images of CD63 (green) and insulin (red) (upper panel), and CD63 (green) and glucagon (red) (lower panel) in pancreatic islets (Scale bars, 50 μm). (N = 3 independent experiments). b, Violin plots representing expression of Cd63 across different pancreatic islet cell types from mice fed with RD and HFD. Minimum to maximum values are shown by the whiskers, the bounds of boxes represent the first and third quartiles and the center line indicates the median. α cells [RD = 802 cells, HFD = 430 cells]; β cells [RD = 4514 cells, HFD = 3050 cells]; γ cells [RD = 182 cells, HFD = 130 cells]; δ cells [RD = 461 cells, HFD = 377 cells]; Endothelial cells [RD = 196 cells, HFD 140 cells]; Epithelial cells [RD = 60 cells, HFD = 79 cells]; Immune cells [RD = 69 cells, HFD = 92 cells]. c, Representative immunofluorescence images showing CD63 (green) and insulin (red) in the different portions of the pancreas (head, body and tail) from 10-week old WT male mice (left panel) (Scale bars, 50 μm). Quantification of the percentage of β cells that are CD63hi in the different parts of the pancreas (right panel) (N = 4 mice per group). Data are presented as mean ± S.E.M. Two-way ANOVA is used for comparison of multiple groups. d, Scatterplots showing the sequential gating scheme to determine CD63 protein expression by flow cytometry in β cells from mouse insulin promoter-GFP transgenic mice. Results are representative of 3 independent experiments. e, Scatterplots showing the sequential gating protocol to isolate CD63hi and CD63lo β cells from WT mice by FACS (in total 60057 CD63hi and 60064 CD63lo β cells). Source numerical data are available in source data.
Extended Data Fig. 4 Molecular characterization of CD63hi β cells.
a, Venn diagram illustrating the overlap of differentially expressed genes (DEGs) obtained by bulk RNA-seq and genes enriched in macrophages (left panel). Scatterplot of RNA-seq analysis from FAC-sorted CD63hi and CD63lo β cells. DEGs with a false discovery rate (FDR) < 0.05 are in green and those that are not differentially expressed are in grey (right panel). b, Heatmap of 396 DEGs from FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-seq. (N = 2, each replicate is pooled from 5 mice). c, Heatmap showing the enriched pathways in β cell clusters 0-3 and CD63hi and CD63lo β cells. d, Differentially expressed transcription factors (DETs) in both CD63hi and CD63lo β cells (p-value was calculated using two-tailed Benjamini-Hochberg). e, Top 20 transcription factors predicted by upstream pathway analysis in CD63hi β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). f, Statistical significance of the predicted transcription factors from CD63hi β cells (e) in Cluster 1 β cells (p-value was calculated using two-tailed Benjamini-Hochberg). Source numerical data are available in source data.
Extended Data Fig. 5 CD63hi β cell maturation.
a, Volcano plot showing differentially expressed genes (DEGs) in immature and mature β cells52. b, Gene expression of Cd63, Ins1, Cd81 and Rbp4 by RNA-seq on FAC-sorted β cells during maturation. Gene structure and chromosome number are indicated for each panel53. Data are publicly available at Huising Lab website (https://www.huisinglab.com/). c, Volcano plot showing differentially expressed genes (DEGs) in embryonic and mature β cells54. d, Gene expression of Cd63 and Ins1 by RNA-seq on FAC-sorted β cells during perinatal maturation. Gene structure and chromosome number are indicated for each panel53. Data are publicly available at Huising Lab website (https://www.huisinglab.com/).
Extended Data Fig. 6 Functional analysis of CD63hi β cells.
a, Contour plots of mitochondrial membrane potential in β cells incubated with 2.8 or 20 mM glucose. Histogram and quantification of CD63 expression in β cells with low and high mitochondrial membrane potential after incubation with 20 mM glucose. Results are representative of 4 independent experiments. Data are presented as mean ± S.E.M. Paired two-tailed t test is used for comparison. b, NAD(P)H fluorescent patterns of CD63hi and CD63lo β cells after incubation with 20 mM glucose. Quantification of NAD(P)H levels after incubation with 2.8 or 20 mM glucose (N = 3 independent experiments per group). Paired two-tailed t test is used for comparison. c, Analysis and quantification of cellular granularity in CD63hi and CD63lo β cells assessed by side scatter (SSC). (N = 4 independent experiments per group). Paired two-tailed t test is used for comparison. d, Scatterplots and quantification showing the distribution of CD63hi and CD63lo β cells in the different subpopulations of mouse insulin promoter-GFP transgenic β cells (N = 3 independent experiments per group; 5 mice per experiment). Welch’s unpaired two-tailed t test is used for comparison. e, Representative immunofluorescence images of pseudo-islets from mice showing β cells (Insulin, Red) and α cells (Glucagon, Green) (Scale bars, 50 μm). N = 5 pseudo-islets per group. f, Static glucose-stimulated insulin secretion (GSIS) assay normalized by insulin content in CD63hi and CD63lo murine pseudo-islets. Data are pooled from 4 independent experiments. Paired two-tailed t test is used for comparison. g, Insulin content of CD63hi and CD63lo murine pseudo-islets normalized by intracellular protein (N = 5 independent experiments per group). Paired two-tailed t test is used for comparison. h, Representative immunofluorescence images of murine pseudo-islets showing Insulin (Red), CD63 (Green) and GADPH (Blue) (Scale bars, 50 μm). N = 5 pseudo-islets per group. Data are presented as mean ± S.E.M. Source numerical data are available in source data.
Extended Data Fig. 7 Insulin secretion of CD63hi and CD63lo β cells in response to GLP-1 agonist.
a, Flow cytometry analysis for annexin V and 7-AAD in isolated CD63hi and CD63lo β cells from WT mice. Data are representative of 3 independent experiments (cells pooled from at least 5 mice per experiment). b, Static glucose-stimulated insulin secretion (GSIS) assay in the absence or presence of 100 μM 3-isobutyl-1-methylxanthine (IBMX). Results are normalized by insulin content in CD63hi and CD63lo murine pseudo-islets (N = 3 per group). Welch’s unpaired two-tailed t test is used for comparison. c, Venn diagram illustrating the overlap of differentially expressed genes (FDR < 0.01) between Cluster 0 β cells obtained by scRNA-seq and sorted CD63lo β cells determined through bulk RNA-seq. Biological process enrichment analysis of differentially expressed genes in both Cluster 0 and CD63lo β cells. d, Glp1r mRNA expression in sorted CD63hi and CD63lo mouse β cells determined by bulk RNA-seq. Comparisons were performed by Wald test. Benjamini–Hochberg corrected two-tiled p-value. e, Static GSIS assay in the absence or presence of 5 nM Exendin-4 (Ex4). Results are normalized by insulin content in CD63hi and CD63lo murine pseudo-islets (N = 4 replicates per group). Results are representative of 3 independent experiments. Data are presented as mean ± S.E.M. Paired two-tailed t test is used for comparison. Source numerical data are available in source data.
Extended Data Fig. 8 Molecular analysis of human CD63hi β cells.
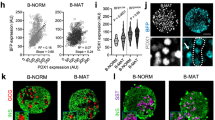
a, Quantification of the percentage of human β cells that are CD63hi (Fig. 3a). N = 3 non-diabetic human donors. Data are presented as mean ± S.E.M. b, UMAP showing the clustering of cells by constructing a Shared Nearest Neighbor (SNN) graph with resolution of 0.5 integrating cells from non-diabetic (ND) and donors with type 2 diabetes (T2D). c, UMAP indicating cells from ND and T2D donors in Extended Data Fig. 8b. d, Violin plots showing CD63 expression in H0 and H1 β cell subclusters in ND and T2D. e, Box plots showing the pseudo bulked median expression levels of CD63 in H0 and H1 β cell subclusters from ND (N = 22) and T2D (N = 14) donors. Each dot represents the median expression of one donor. Boxes represent the first and third quartiles, center line denotes the median and whiskers showed the maximum and minimum value. f, CD63 mRNA expression in whole islets from male (N = 10) and female (N = 8) ND donors determined by RT-qPCR. g-h, Frequency of Cluster H1 cells (g) and body mass index (BMI) (h) in T2D donors by sex. Donors with at least 13 β cells are included in the analysis. Male (N = 6), Female (N = 8). i, Pathway enrichment analysis of differentially expressed genes (DEGs) in human Cluster H1 β cells. j, Venn diagram illustrating the overlap of DEGs (FDR < 0.01) among Cluster 0 and 1 mouse β cells obtained by scRNA-seq and human H1 subcluster. Pathway enrichment analysis of DEGs by scRNA-seq in Cluster 1 mouse and human H1 β cells. k, Volcano plot showing DEGs in H1 β cells between T2D and ND donors. l, Pathway enrichment analysis of DEGs in H1 β cells from T2D donors. m, Pathway enrichment analysis of DEGs in H1 β cells from ND donors. Data are presented as mean ± S.E.M. and Welch’s unpaired two-tailed t test is used for comparison (f, g, and h), and p-values were calculated using two-tailed Benjamini-Hochberg in I, j, k, l and m. Source numerical data are available in source data.
Extended Data Fig. 9 Effects of obesity on functional and transcriptional profiles of β cells.
a, Representative flow cytometry plot for CD63 and CD81 staining on β cells from regular diet (RD) and high fat diet (HFD) fed male mice. b, Percentage of CD63hi and CD81+ β cells from RD and 6-weeks HFD-fed male mice. N = 5 mice per group. c, Percentage of CD63hi β cells from RD and 6-weeks HFD-fed female mice using flow cytometry. N = 4 mice per group. d, Representative immunofluorescence images showing CD63 (green) and insulin (red) in pancreas from WT female mice fed with RD or HFD for 6 weeks. (Scale bars, 50 μm). e, Quantification of the percentage of β cells that are CD63hi in female mice fed RD or HFD for 6 weeks (N = 4 mice per group). f, Plot showing differentially expressed genes (DEG) between β cells from RD and HFD-fed male mice. g, Violin plots representing expression of Cd63 across different β cell clusters from male mice fed with HFD. Whiskers show the minimum to maximum values, bounds of boxes represent first and third quartiles and the center line indicates the median. Cluster 0 (N = 1281 cells), Cluster 1 (N = 758), Cluster 2 (N = 767), Cluster 3 (N = 244 cells). h–k, Volcano plot showing DEGs between RD- and HFD-fed mice and pathways enriched in Cluster 0 (h), Cluster 1 (i), Cluster 2 (j) and Cluster 3 (k) β cells. l, Venn diagram illustrating the overlap of DEGs (FDR < 0.01) between the bulk β cell population (all β cells from this scRNA-seq study) from mice fed with HFD and β cell population from mice fed with HFD determined through bulk RNA-seq56. Pathway analysis of shared DEGs. m, Glucose stimulated insulin secretion assay from pseudo-islets derived from CD63hi and CD63lo β cells from RD and HFD-fed mice. CD63lo (RD [N = 8 Samples]; HFD [N = 8 Samples) and CD63hi β cells (RD [N = 9 Samples]; HFD (N = 8 Samples). Data are presented as mean ± S.E.M. and P values were determined by Welch’s unpaired two-tailed t test (b, c, e, and m). Source numerical data are available in source data.
Supplementary information
Supplementary Tables 1–6
Supplementary Table 1. Purity assessment of FAC-sorted CD63hi and CD63lo mouse beta cells. Data are presented as mean ± s.e.m. (N = 5 independent experiments). Supplementary Table 2. Donor characteristics for pancreatic section used in the immunofluorescence staining experiments. Supplementary Table 3. Donor characteristics for human islets used in the in vitro experiments. Supplementary Table 4. Purity assessment of FAC-sorted CD63hi and CD63lo human beta cells. Data are presented as mean ± s.e.m. (N = 5 independent experiments). Supplementary Table 5. Donor characteristics for human islets used in Fig. 3j. Supplementary Table 6. qPCR primers.
Source data
Source Data Fig. 1
Numerical source data.
Source Data Fig. 2
Numerical source data.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Numerical source data.
Source Data Fig. 5
Numerical source data.
Source Data Fig. 6
Numerical source data.
Source Data Extended Data Fig./Table 1
Numerical source data.
Source Data Extended Data Fig./Table 2
Numerical source data.
Source Data Extended Data Fig./Table 3
Numerical source data.
Source Data Extended Data Fig./Table 4
Numerical source data.
Source Data Extended Data Fig./Table 5
Numerical source data.
Source Data Extended Data Fig./Table 6
Numerical source data.
Source Data Extended Data Fig./Table 7
Numerical source data.
Source Data Extended Data Fig./Table 8
Numerical source data.
Source Data Extended Data Fig./Table 9
Numerical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rubio-Navarro, A., Gómez-Banoy, N., Stoll, L. et al. A beta cell subset with enhanced insulin secretion and glucose metabolism is reduced in type 2 diabetes. Nat Cell Biol 25, 565–578 (2023). https://doi.org/10.1038/s41556-023-01103-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-023-01103-1
This article is cited by
-
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity
Diabetologia (2024)
-
A Model for Detecting Type 2 Diabetes Using Mixed Single-Cell RNA Sequencing with Optimized Data
SN Computer Science (2023)