Abstract
T cell acute lymphoblastic leukaemia (T-ALL) is an aggressive malignancy with poor prognosis, but a decisive marker and effective treatment for leukaemia stem cells (LSCs) remain unclear. Here, using lineage tracing, limiting dilution assays and in vivo live imaging approaches, we identify rare inhibitory receptor programmed cell death 1 (PD-1)-expressing cells that reside at the apex of leukaemia hierarchy for initiation and relapse in T-ALL. Ablation of PD-1-expressing cells, deletion of PD-1 in T-ALL cells or blockade of PD-1 or PD-1 ligand 1 significantly eradicated LSCs and suppressed disease progression. Combination therapy using PD-1 blockade and chemotherapy substantially extended the survival of mice engrafted with mouse or human T-ALL cells. Mechanistically, PD-1+ LSCs had high NOTCH1–MYC activity for disease initiation. Furthermore, PD-1 signalling maintained quiescence and protected LSCs against T cell receptor-signal-induced apoptosis. Overall, our data highlight the hierarchy of leukaemia by identifying PD-1+ LSCs and provide a therapeutic approach for the elimination of LSCs through PD-1 blockade in T-ALL.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All the sequencing data generated in this study have been deposited into Gene Expression Omnibus under accession number GSE215392. These data are publicly available. Complete clinical data for 261 patients with T-ALL were downloaded from the TARGET dataset (2 March 2020; https://ocg.cancer.gov/programs/target/data-matrix). The data that support the findings of this study are available from the corresponding authors upon reasonable request. Source data are provided.
Code availability
The tools supporting this paper are available on GitHub, including Trim Galore (v.0.6.7), Salmon (v.0.13.1), R (v.3.6.1 or 4.0), python (v.2.7.2 or 3.6.0) and the packages tidyverse (v.1.3.1), DESeq2 (v.1.36.0), ggplot2 (v.3.3.6), pheatmap (v.1.0.12), clusterProfiler (v.4.4.4) and GSEA (https://www.gsea-msigdb.org/gsea/index.jsp).
References
Terwilliger, T. & Abdul-Hay, M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 7, e577 (2017).
Belver, L. & Ferrando, A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 16, 494–507 (2016).
Pui, C. H., Robison, L. L. & Look, A. T. Acute lymphoblastic leukaemia. Lancet 371, 1030–1043 (2008).
Van Vlierberghe, P. & Ferrando, A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 122, 3398–3406 (2012).
Cox, C. V. et al. Characterization of a progenitor cell population in childhood T-cell acute lymphoblastic leukemia. Blood 109, 674–682 (2007).
Armstrong, F. et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood 113, 1730–1740 (2009).
Chiu, P. P., Jiang, H. & Dick, J. E. Leukemia-initiating cells in human T-lymphoblastic leukemia exhibit glucocorticoid resistance. Blood 116, 5268–5279 (2010).
Guo, W. et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 453, 529–533 (2008).
McCormack, M. P. et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879–883 (2010).
Giambra, V. et al. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-θ and reactive oxygen species. Nat. Med. 18, 1693–1698 (2012).
Dong, H. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002).
Iwai, Y. et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl Acad. Sci. USA 99, 12293–12297 (2002).
Zou, W., Wolchok, J. D. & Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv324 (2016).
Zou, W. & Chen, L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 8, 467–477 (2008).
Page, D. B., Postow, M. A., Callahan, M. K., Allison, J. P. & Wolchok, J. D. Immune modulation in cancer with antibodies. Annu. Rev. Med. 65, 185–202 (2014).
Asnafi, V. et al. Age-related phenotypic and oncogenic differences in T-cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood 104, 4173–4180 (2004).
Pear, W. S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996).
Herranz, D. et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 20, 1130–1137 (2014).
Keir, M. E., Butte, M. J., Freeman, G. J. & Sharpe, A. H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 (2008).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Weng, A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Medyouf, H. et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood 115, 1175–1184 (2010).
Sanda, T. et al. Interconnecting molecular pathways in the pathogenesis and drug sensitivity of T-cell acute lymphoblastic leukemia. Blood 115, 1735–1745 (2010).
Paczulla, A. M. et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 572, 254–259 (2019).
Kikushige, Y. et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell 7, 708–717 (2010).
Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994).
Radtke, F., Wilson, A., Mancini, S. J. & MacDonald, H. R. Notch regulation of lymphocyte development and function. Nat. Immunol. 5, 247–253 (2004).
King, B. et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013).
Palomero, T. et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA 103, 18261–18266 (2006).
Mathieu, M., Cotta-Grand, N., Daudelin, J. F., Thébault, P. & Labrecque, N. Notch signaling regulates PD-1 expression during CD8+ T-cell activation. Immunol. Cell Biol. 91, 82–88 (2013).
Langerak, A. W. et al. Molecular and flow cytometric analysis of the Vβ repertoire for clonality assessment in mature TCRαβ T-cell proliferations. Blood 98, 165–173 (2001).
Trinquand, A. et al. Triggering the TCR developmental checkpoint activates a therapeutically targetable tumor suppressive pathway in T-cell leukemia. Cancer Discov. 6, 972–985 (2016).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Vilimas, T. et al. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 13, 70–77 (2007).
Kourtis, N. et al. Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat. Med. 24, 1157–1166 (2018).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019).
Utzschneider, D. T. et al. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J. Exp. Med. 213, 1819–1834 (2016).
Sheppard, K. A. et al. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3ζ signalosome and downstream signaling to PKCθ. FEBS Lett. 574, 37–41 (2004).
Pollyea, D. A. & Jordan, C. T. Therapeutic targeting of acute myeloid leukemia stem cells. Blood 129, 1627–1635 (2017).
Jones, C. L., Inguva, A. & Jordan, C. T. Targeting energy metabolism in cancer stem cells: progress and challenges in leukemia and solid tumors. Cell Stem Cell 28, 378–393 (2021).
Wartewig, T. et al. PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature 552, 121–125 (2017).
Casulo, C. et al. T-cell lymphoma: recent advances in characterization and new opportunities for treatment. J. Natl Cancer Inst. 109, djw248 (2017).
Ratner, L., Waldmann, T. A., Janakiram, M. & Brammer, J. E. Rapid progression of adult T-cell leukemia-lymphoma after PD-1 inhibitor therapy. N. Engl. J. Med. 378, 1947–1948 (2018).
Rauch, D. A. et al. Rapid progression of adult T-cell leukemia/lymphoma as tumor-infiltrating Tregs after PD-1 blockade. Blood 134, 1406–1414 (2019).
Gao, J. et al. Therapeutic targeting of Notch signaling and immune checkpoint blockade in a spontaneous, genetically heterogeneous mouse model of T-cell acute lymphoblastic leukemia. Dis. Models Mech. 12, dmm040931 (2019).
Venezia, T. A. et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2, e301 (2004).
Lerner, C. & Harrison, D. E. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol. 18, 114–118 (1990).
Medyouf, H. et al. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat. Med. 13, 736–741 (2007).
Hanke, T. et al. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity 11, 67–77 (1999).
Eppert, K. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17, 1086–1093 (2011).
Gal, H. et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia 20, 2147–2154 (2006).
Holleman, A. et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 351, 533–542 (2004).
Perry, J. M. et al. Overcoming Wnt–β-catenin dependent anticancer therapy resistance in leukaemia stem cells. Nat. Cell Biol. 22, 689–700 (2020).
Grey, W. et al. CKS1 inhibition depletes leukemic stem cells and protects healthy hematopoietic stem cells in acute myeloid leukemia. Sci. Transl. Med. 14, eabn3248 (2022).
Acknowledgements
We thank staff at the core facilities for medical science at Zhongshan School of Medicine, Sun Yat-sen University for technical support, and H. Wu (Beijing University) and W. Guo (Zhejiang University) for providing Pten-null mice. We thank the National Key Research and Development Program of China (2018YFA0107200 to M.Z.), the National Natural Science Foundation of China (82170112 to M.Z.), Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y485 to L.J.), Sanming Project of Medicine in Shenzhen (SZSM201911004 to M.Z.), Advanced Medical Technology Center, The First Affiliated Hospital, Zhongshan School of Medicine and Sun Yat-sen University to M.Z. for generous support. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
X.X. and W. Zhang designed and performed most of the experiments and analysed the data. L.X., Minyi Zhao, J.W., Y.Q. and Q.L. contributed to experiments related to clinical samples. Y.Y., W.M., C.H., S.X., X.H., F.T. and J.N. contributed to T-ALL animal experiments. W. Zheng contributed to Pten-null T-ALL. J.M.P. and L.J. contributed to scientific discussion and manuscript preparation. M.Z. supervised the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Iannis Aifantis, Weiyi Peng and Toshio Suda for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of PD-1+ T-ALL cells.
Schematic depiction (top) and the representative FCAS plots (bottom) for the NOTCH1-induced T-ALL model. Hematopoietic stem and progenitor cells (HSPCs, Lin–Sca1+C-Kit+ cells) were enriched at d5.5 after 5FU treatment. HSPCs were infected with the NOTCH1 (co-expressed with GFP) virus, and the GFP+ T-ALL cells were analyzed after infection, and in the bone marrow of recipients at indicated days after transplantation. Plots are representative of n = 3 biological repeats.
Extended Data Fig. 2 Characterization of Pd1-2A-CreERT2; TdTomato reporter mice.
a, Schematic diagram of Pd1-2A-CreERT2 mouse line. The homologous recombination PCR results of F1 generation heterozygous mice. The numbers indicated individual mice; wt, wild type; M, 1 kb DNA ladder. b, Schematic diagram and the Southern Blot result of homologous recombination. The numbers indicated individual mice. c, Schematic diagram of subcutaneous transplantation of B16 melanoma cells into Pd-1-CreERT2; tdTomato or Pd-1–/– mice. d,e, Six weeks after injection, the tumor tissues were digested and FACS analyzed at one day after tamoxifen injection for Pd-1-CreERT2; tdTomato mice (d) and Pd-1–/– mice (e). Plots are representative of n = 3 biological repeats. f, Schematic depicting strategy of T cells with anti-CD3/CD28 antibodies and 4-hydroxytamoxifen for 72 h under in vitro culture conditions. g,h, Representative FACS plots of PD-1 and Tomato expression in CD3+ T cells from Pd-1-CreERT2; tdTomato mice (g) or Pd-1–/– mice (h) with indicated treatments. Plots are representative of n = 3 independent repeats. i, Quantification of GFP+ T-ALL cells in peripheral blood at indicated time points after transplantation. (n = 5 mice). j, The survival curves of T-ALL mice (n = 5 mice). k, Representative H&E staining images for the spleen, liver, and lung sections from control mice or T-ALL mice at the indicated weeks after transplantation. Scale bar 50 µm. l, Representative FACS plots of PD-1 expression in GFP+ T-ALL cells in peripheral blood of T-ALL mice at d42 after transplantation. k,l, Images and plots are representative of n = 5 mice. All data are shown as mean ± s.d.
Extended Data Fig. 3 PD-1+ LSCs in Pten null T-ALL and NOTCH1-induced T-ALL.
a, PD-1 expression in Pten null T-ALL cells (SSClowCD45high population) in the bone marrow. b, The frequency of SSClowCD45high leukemic cells in peripheral blood of recipients engrafted with indicated numbers of PD-1+ and PD-1– cells. (n = 5 mice for each group). c, The survival curves of recipients engrafted with indicated numbers of PD-1– or PD-1+ Pten null T-ALL cells (n = 5 mice for each group). d, Competitive repopulating units (CRU) analysis for PD-1+ or PD-1– Pten null T-ALL cells in recipients (n = 5 mice for each group). e, PD-1– cells did not generate PD-1+ cells in NOTCH1-induced T-ALL recipients at 6 weeks after transplantation when the leukemic chimera rate reached 90% in the bone marrow. f,g, Quantification of NOTCH1-induced T-ALL cells in peripheral blood at indicated time points after transplantation (f) and the survival curve (g) of recipients engrafted with purified PD-1+ or PD-1– T-ALL cells from secondary recipients at 3 weeks after transplantation. (n = 5 mice for each group). h,i, Schematic depicting the sorting and transplantation strategy for PD-1+ and PD-1– T-ALL cells from Pten null T-ALL. The representative FACS plots (h) and quantification (i) of PD-1+ and Lin–CD3+C-KitMid cells in SSClowCD45high leukemic cells in the bone marrow of B-NDG recipients at 10 weeks after transplantation with PD-1+ or PD-1– Pten null T-ALL cells (n = 5 mice for each group). The P values in b,f,i, were determined by the unpaired two-tailed Student’s t-test; g, was determined by Kaplan–Meier survival analysis; d, was determined by the ELDA platform. Data are shown as mean values ± s.d.
Extended Data Fig. 4 PD-1 is not required for normal HSCs or cell-nonautonomously required for T-ALL progress.
a-d, The absolute numbers (a), cell cycle (b), apoptosis rate (c), and colony-forming ability (d) of HSCs in the bone marrow of Pd-1 deleted mice and littermate controls. (n = 4 mice for each group). e-g, Schematic strategy (e) and quantification of HSC function by transplantation assay. 500 HSCs from control or Pd-1 deleted mice (CD45.2+) were transplanted into irradiated mice (CD45.1+) along with 2 × 105 recipient bone marrow cells (CD45.1+). PB analysis for total engrafted donor cells at the indicated number of weeks after transplantation (f) and the percentage of donor-derived B, T, and myeloid lineage cells at 16 weeks after transplantation (g) (n = 8 mice for each group). h,i, Schematic strategy for T-ALL transplantation using control or Pd-1 deleted recipients (h) and quantification of T-ALL cells in peripheral blood in control or Pd-1 deleted recipients at the indicated time after transplantation with T-ALL cells (i) (n = 6 mice for each group). j, Survival curves of Pd1+/+ or Pd-1–/– recipients engrafted with T-ALL cells (n = 6 mice for each group). The P values in a, b, c, d, g, i, were determined by the unpaired two-tailed Student’s t-test. The P value in j was determined by Kaplan–Meier survival analysis. ns, not significant. All data are shown as mean ± s.d.
Extended Data Fig. 5 Characterization of PD-1 expression and PD-1+ cells in human T-ALL.
a, Diagram of the correlation between PD-1 expression and disease prognosis. white blood cell count (WBC), minimal residual disease (MRD), chemotherapy (chemo). b, Survival curves of T-ALL patients with high PD-1 (n = 130 patients) or low PD-1 expression (n = 131 patients). c, Representative FACS plots of PD-1 and CD34 expression in primary human T-ALL cells from the bone marrow aspirations from T-ALL patients (left), and human CD45+CD3+ T-ALL cells in peripheral blood of B-NDG recipients at 8 weeks after transplantation with 1 × 107 human T-ALL cells (right). Successfully engrafted T-ALL PDX experiments (7 out of 15 patients) were presented. d,e, Representative FCAS plots (d) and quantification (e) of human CD45+CD3+PD-1+ T-ALL cells in the bone marrow of B-NDG recipients at 8 weeks after transplantation. f, The Wright-Giemsa staining of peripheral blood, H&E staining of spleen, lung, liver, and anti-human CD45 staining of bone marrow section from B-NDG mice at d14 after T-ALL cell transplantation. Similar images were obtained in three independent experiments. Two individual T-ALL patients were presented. Peripheral blood and bone marrow section, scale bar 10 µm; spleen, lung, and liver, scale bar 50 µm. g,h, Representative images of Wright-Giemsa staining (g) and quantification of cell-to-nucleus size ratio (h) of human PD-1– and PD-1+ T-ALL cells (n = 50 cells from 3 patients). Scale bar 5 µm. i, Representative FACS plots of PD-1– and PD-1+ human T-ALL cells. Similar plots were obtained in three independent patients. The P values were determined by pearson correlation analysis (a), Kaplan–Meier survival analysis (b), the paired two-tailed Student’s t-test (e), and the unpaired two-tailed Student’s t-test (h). All data are shown as mean ± s.d.
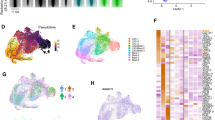
Extended Data Fig. 6 Transcriptional Characterization of PD-1+ LSCs in Notch1-induced T-ALL and Pten null T-ALL.
a,b, Heatmap and expression score for indicated gene sets in PD-1– and PD-1+ T-ALL cells (n = 3 mice). Leukemia stem cell (LSC), acute myeloid leukemia (AML), and acute lymphoid leukemia (ALL). c, Schematic depicting strategy for the transcriptional analysis for PD-1+ and PD-1– leukemic cells from Pten null T-ALL mice. d, GO term analysis of PD-1+ T-ALL cells of differentially expressed genes in PD-1– and PD-1+ Pten null T-ALL cells (n = 3 mice). e, Heatmap of differentially expressed genes in PD-1– and PD-1+ Pten null T-ALL cells (n = 3 mice). f,g, GSEA (top) and analysis (bottom) of the cell cycle (f) and ROS (g) of PD-1– and PD-1+ Pten null T-ALL cells (n = 3 mice). h-j, GSEA of the apoptotic pathway (h), early T lymphocyte genes, MYC pathway, and MYC upregulated genes (i), and NOTCH1 signal pathway (j) in PD-1– and PD-1+ Pten null T-ALL cells (n = 3 mice). k,l, Heatmap of differentially expressed genes and expression score for LSC genes in AML (k) and chemoresistant genes in ALL (l) in PD-1– and PD-1+ Pten null T-ALL cells (n = 3 mice). The P values in a(right),b (right),f(bottom),k(right),l(right) were determined by the unpaired two-tailed Student’s t-test. All data are shown as mean ± s.d.
Extended Data Fig. 7 Transcriptional Characterization of PD-1+ LSCs in human T-ALL.
a, Schematic depicting strategy for the transcriptional analysis for PD-1+ and PD-1– leukemic cells from human T-ALL. b, GO term analysis of PD-1+ T-ALL cells of differentially expressed genes in PD-1– and PD-1+ human T-ALL cells. (n = 3 individual patients). c, Heatmap of differentially expressed genes in PD-1– and PD-1+ human T-ALL cells (n = 3 individual patients). d,e, GSEA (top) and analysis (bottom) of cell cycle (d) and ROS (e) of PD-1– and PD-1+ human T-ALL cells (n = 3 individual patients). f-h, GSEA of the apoptotic pathway (f), early T lymphocyte genes, MYC pathway, and MYC upregulated genes (g), and NOTCH1 signal pathway (h) in PD-1– and PD-1+ human T-ALL cells (n = 3 individual patients). i,j, Heatmap of differentially expressed genes and expression score for leukemia stem cell (LSC) genes in acute myeloid leukemia (AML) (i) and chemoresistant genes in acute lymphoid leukemia (ALL) (j) in PD-1– and PD-1+ human T-ALL cells (n = 3 individual patients). The P values in d(bottom),i(right),j(right), were determined by the unpaired two-tailed Student’s t-test. All data are shown as mean ± s.d.
Extended Data Fig. 8 Characterization of surface markers and NOTCH1 signaling in PD-1+ LSCs.
a,b, Quantification of T-ALL cells in peripheral blood (a) and survival curve (b) of recipients engrafted with indicated T-ALL cells (n = 5 mice). c, The expression, and quantification of indicated LSC markers (n = 4 mice). d, The survival curve of mice received the indicated numbers of T-ALL cells (n = 5 mice). e,f, Summary of limiting dilution experiments (f) and the competitive repopulating units (f). g, PD-1 expressioin in Lin–CD3+c-KitMid T-ALL cells in Pten null T-ALL. h, FACS analysis showing Lin, CD3, and c-Kit expression in PD-1+ and PD-1– Pten null T-ALL cells. i, The survival curve of T-ALL mice received the indicated T-ALL cells (n = 5 mice). j,k, Summary of limiting dilution experiments (j), and the competitive repopulating units (k). l, The expression, and quantification of indicated LSC markers in human T-ALL cells from recipients (n = 5 individual patients). m,n, Quantification of MYC-regulated gene expression in T-ALL cells (n = 3 mice). o, Quantification of indicated proteins (n = 3 mice) (Related to Fig. 5l). p,q, Western blots for T-ALL cells from Pten null T-ALL (p) and human T-ALL (q). Three individual mice or human samples were presented. r,s, Quantification of MYC regulated genes in human T-ALL cells (n = 3 individual patients). t, The NOTCH1 mutation rate in human T-ALL cells (n = 7 individual patients). u,v, Western blots (u) and correlation analysis (v) for indicated proteins in human T-ALL cells. 12 individual patient samples were presented. w, Relative PD-1 mRNA levels in indicated cells with NOTCH1 inhibitor (LY411575) treatment for 72 h (n = 3 mice). β-actin was used as a loading control (p,q,u). The P values were determined by the unpaired two-tailed Student’s t-test (a,c,l,m,n,o,r,s,t,w), Kaplan–Meier survival analysis (b,i), the ELDA platform (f,k), and pearson correlation analysis (v). All data are shown as mean ± s.d.
Extended Data Fig. 9 PD-1 signaling protects PD-1+ LSCs from TCR-induced apoptosis.
a, Quantification of pERK expression in PD-1– and PD-1+ T-ALL cells after TCR stimulation (n = 3 mice) (Related to Fig. 6e). b, Representative FACS plots for TCR expression in PD-1+ and PD1– T-ALL cells (n = 3 mice). c, Quantification of Western blots for indicated proteins in PD-1+ and PD-1– T-ALL cells (n = 3 mice) (Related to Fig. 6g). d, Western blots of indicated proteins in PD-1– and PD-1+ T-ALL cells with SHP-1 inhibitor (SHP-1i), SHP-2 inhibitor (SHP-2i) or combination (SHP-1/2i) for 72 h (n = 3 mice). e-g, Cell cycle analysis (e), proliferation (f), and apoptosis (g) of PD-1– and PD-1+ T-ALL cells with indicated treatment for 72 h (e, g) or indicated time (f) (n = 3 mice). h, Schematic diagram for PD-1– and PD-1+ T-ALL cell purification, and control (Ctrl) or PD-1 overexpression (OE) virus infection. i-k, Representative FACS plots (i), western blots and quantification of MYC and NOTCH1 proteins (j), and the apoptosis analysis (k) for PD-1+ and PD-1– T-ALL cells with PD-1 overexpression as indicated (n = 4 mice). Three individual mice were presented (j). l,m, The frequency of GFP+ T-ALL cells in peripheral blood at the indicated time after transplantation (l), and the survival curves (m) of indicated recipients (n = 5 mice). n, Western blots (left) and quantification (right) of indicated proteins in human PD-1+ and PD-1– T-ALL cells treated with peptide MHC (pMHC) (n = 3 mice). The P values in a,c,e,g,n(right) were determined by the unpaired two-tailed Student’s t-test. The P values in f, j(right),k,l were determined by a two-way analysis of variance (ANOVA). The P values in m were determined by Kaplan–Meier survival analysis. All data are shown as mean ± s.d.
Extended Data Fig. 10 The expression and role of PD-L1 for T-ALL.
a,b, Representative imaging of GFP+Tomato+ (PD-1+ cells), GFP+Tomato– (PD-1–) T-ALL cells, and PD-L1+ cells were analyzed at d1 after tamoxifen injection in T-ALL mice engrafted with Pd-1-CreERT2; tdTomato T-ALL cells at 3 weeks after transplantation. The yellow arrows indicated representative PD-L1+ T-ALL cells (GFP+), PD-L1+ normal T cells (GFP–CD3+), PD-L1+ myeloid cells (CD11b+), PD-L1+ endothelial cells (ECs, Endomucin+) (a), and PD-1+ T-ALL cells (Tomato+GFP+) (b). The scale bar was 5 µm. c, Quantification of the distance between PD-1–, PD-1+ T-ALL cells, and randomly placed spots with PD-L1 expressing cells in the bone marrow of T-ALL mice cells at 3 weeks after transplantation (n = 3 mice). d, Quantification for Fig. 6l (d) (n = 3 mice). e, Schematic depicting strategy of using anti-CD3/CD28 antibodies to stimulate TCR signaling in Pd-l1 deleted PD-1+ and PD-1– T-ALL cells (left) and western blots of indicated proteins in Pd-l1 deleted PD-1– or PD-1+ T-ALL cells with indicated treatments for 72 h (right). Similar blots were obtained in three independent experiments. f, Quantification for Fig. 6o (n = 3 mice). The P values in c,d,f were determined by a two-way analysis of variance (ANOVA). All data are shown as mean ± s.d.
Supplementary information
Supplementary Tables
Supplementary Table 1: Summary of limiting dilution experiments for PD-1– and PD-1+ T-ALL cells. The mice that died from leukaemia in each group and the inverse of LSC frequency as well as upper and lower estimates are provided. Supplementary Table 2: Characteristics of patients with T-ALL. Detailed information for 15 patients with T-ALL in the PDX experiments is provided. Supplementary Table 3: qPCR primer list. The qPCR primers for mRNA expression experiments and chromatin immunoprecipitation assay are provided. Supplementary Table 4: Gene sets for bioinformatics analysis. The gene lists in each gene set, including cell cycle, apoptosis, response to oxidation stress, NOTCH1 signal, PI3K–AKT pathway, MYC pathway, MYC target, early T lymphocyte, leukaemia stemness genes, AML LSC genes, drug resistance and the negative select genes are provided.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, X., Zhang, W., Xuan, L. et al. PD-1 signalling defines and protects leukaemic stem cells from T cell receptor-induced cell death in T cell acute lymphoblastic leukaemia. Nat Cell Biol 25, 170–182 (2023). https://doi.org/10.1038/s41556-022-01050-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-01050-3
This article is cited by
-
Revisiting PD-1 to target leukaemic stem cells
Nature Cell Biology (2023)
-
Targeted therapy and immunotherapy for T cell acute lymphoblastic leukemia/lymphoma
Annals of Hematology (2023)