Abstract
The biological purpose of long non-coding RNAs (lncRNAs) is poorly understood. Haploinsufficient mutations in HNF1A homeobox A (HNF1A), encoding a homeodomain transcription factor, cause diabetes mellitus. Here, we examine HASTER, the promoter of an lncRNA antisense to HNF1A. Using mouse and human models, we show that HASTER maintains cell-specific physiological HNF1A concentrations through positive and negative feedback loops. Pancreatic β cells from Haster mutant mice consequently showed variegated HNF1A silencing or overexpression, resulting in hyperglycaemia. HASTER-dependent negative feedback was essential to prevent HNF1A binding to inappropriate genomic regions. We demonstrate that the HASTER promoter DNA, rather than the lncRNA, modulates HNF1A promoter–enhancer interactions in cis and thereby regulates HNF1A transcription. Our studies expose a cis-regulatory element that is unlike classic enhancers or silencers, it stabilizes the transcription of its target gene and ensures the fidelity of a cell-specific transcription factor program. They also show that disruption of a mammalian lncRNA promoter can cause diabetes mellitus.
Similar content being viewed by others
Main
The transcription of genes is controlled by cis-acting promoter and enhancer sequences, many of which harbour disease variants. Mammalian genomes also contain >20,000 long non-coding RNAs (lncRNAs)1,2. Although the function of most lncRNAs has not been explored, some lncRNAs are known to regulate gene transcription3,4. A considerable number of lncRNAs are transcribed from evolutionarily conserved promoters located near genes encoding lineage-specific regulators3,5,6,7, suggesting a cis-regulatory function. For some lncRNAs, knockdown experiments have revealed transcriptional effects on nearby genes8,9,10, while genetic studies have demonstrated bona fide cis-regulatory functions of selected lncRNAs3,11,12,13,14,15,16. There are nevertheless still major gaps in our understanding of the regulatory purpose of cis-acting lncRNAs and how they are fundamentally different from more established gene regulatory elements. Furthermore, the extent to which genetic disruption of cis-regulatory lncRNAs can lead to physiologically relevant phenotypes is unclear.
In this study, we examined HASTER, the promoter of an lncRNA at the HNF1A homeobox A (HNF1A) locus. Mutations in HNF1A, encoding a homeodomain transcription factor17, cause maturity-onset diabetes of the young type 3, the most frequent form of monogenic diabetes mellitus18, while rare and common variants predispose to type 2 diabetes19,20. Studies of homozygous Hnf1a null mutant mice have shown that HNF1A is essential for differentiated cell programs in various organs, whereas human HNF1A haploinsufficiency causes diabetes due to selective abnormalities in pancreatic β cells, indicating that the gene dosage sensitivity of HNF1A is cell specific18,21,22,23,24,25,26. We now show that HASTER is a cell-specific cis-acting transcriptional stabilizer of HNF1A and demonstrate that disruption of this function causes diabetes mellitus in mice.
Results
Evolutionarily conserved co-expression of HNF1A and HASTER
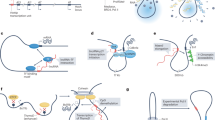
HNF1A-AS1, or Hnf1a-os1 and Hnf1a-os2 in mice, is a putative non-coding transcript that is transcribed from intron 1 of HNF1A and runs in antisense configuration (Fig. 1a). In the present study, we focus on the regulatory function of the promoter of HNF1A antisense transcripts. We named this DNA region HASTER (HNF1A stabilizer). HNF1A antisense transcripts, which we refer to as HASTER RNAs, have previously been proposed to exert trans-regulation of proliferation in cell-based models14,27,28,29,30,31, but so far the transcriptional cis-regulatory function of the lncRNA or its promoter have not been characterized with genetic tools.
a, Human islet RNA-seq (reads per kilobase per million reads, RPKM) and CAGE (normalized tag counts, TPM) showing overlapping and divergent transcription of HNF1A and HASTER (representative examples from four biological replicates). HASTER isoforms were detected by 3′ RACE from human islets. b, Liver strand-specific RNA-seq (RPKM) and Multiz alignments in the indicated species. c, Single-molecule fluorescence in situ hybridization for HASTER (exonic probes) and HNF1A nascent transcripts (intronic probes) in EndoC-βH3 β cells. The yellow arrows indicate co-localization of HASTER and nascent HNF1A transcripts. Quantifications are shown in Extended Data Fig. 2. Scale bar, 2 µm.
We used cap analysis gene expression sequencing (CAGE-seq), RNA sequencing (RNA-seq) and 3′ rapid amplification of complementary DNA ends (RACE) to show that HASTER transcribes myriad transcript isoforms that originate from a major upstream transcriptional start site in human islets and an additional downstream start site in other tissues (Fig. 1a and Extended Data Fig. 1a). Both transcriptional start sites are located in evolutionarily conserved sequences that show active promoter chromatin (high H3K4me3 and low H3K4me1) in islets and liver (Fig. 1b and Extended Data Fig. 1a). HASTER is expressed exclusively in HNF1A-expressing tissues, including the liver, gut, pancreas and kidney, and has the same antisense configuration across species (Fig. 1b and Extended Data Fig. 1b,c). Subcellular fractionation of EndoC-βH3 human β cells showed that HASTER transcripts were associated with chromatin, and single-molecule fluorescence in situ hybridization showed that HASTER transcripts were exclusively present in one or two nuclear foci that co-localized with HNF1A nascent transcripts (Fig. 1c and Extended Data Fig. 2a–c). Therefore, HASTER transcribes an evolutionarily conserved nuclear lncRNA that is co-expressed with HNF1A across tissues.
HASTER is a negative regulator of HNF1A
To study HASTER function, we created a 320-base-pair (bp) deletion of the main HASTER promoter (P1) in human embryonic stem cells (hESCs) (Fig. 2a) and differentiated them into hepatocyte-like cells32. In control cells, HASTER transcripts were already detected at maximal levels at the hepatoblast stage, while HNF1A messenger RNA (mRNA) increased gradually during maturation to hepatocytes (Fig. 2b). HASTER-deleted cells showed increased hepatocyte HNF1A mRNA (mean = 1.3- and 1.6-fold versus control cells for two independent deletions; P = 0.01 and P = 0.04, respectively; Student’s t-test) (Fig. 2b). Thus, HASTER exerts negative regulation of HNF1A in an in vitro human liver cell model.
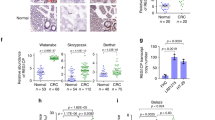
a, Homozygous deletions of the HASTER promoter (two deletions with independent sgRNA pairs) or control deletions in HNF1A intron 1 or AAVS1 were generated in hESCs. b, HNF1A mRNA was increased in differentiated hepatocytes from HASTER mutant hESCs (n = 3 independent clones per deletion). The bar graphs show RPLP0-normalized expression values (means ± s.d.). Statistical significance was determined by two-tailed Student’s t-test. Act. A, Activin A; BMP-4, bone morphogenetic protein 4; HGF, hepatocyte growth factor; OSM, Oncostatin M. c, Schematic of the mouse Hasterf allele. d, Liver RNA levels in seven HasterLKO and eight control mice. The data represent Tbp-normalized values (means ± s.d.). Statistical significance was determined by two-tailed Student’s t-test. e, Liver HNF1A immunofluorescence in the indicated genotypes. Scale bar, 50 µm. f, Western blot for HNF1A on liver extracts (n = 3 mice for each genotype). The bars represent relative expression levels (means ± s.d.). Statistical significance was determined by two-tailed Student’s t-test. Ctrl, control. g,h, Haster was decreased in Hnf1a−/− islets (g; n = 4 Hnf1a−/− and n = 5 Hnf1a+/+ mice) and liver (h; n = 4 mice per genotype). The bars represent relative expression levels (means ± s.d.). Statistical significance was determined by two-sided Wald test with adjusted P values. i, EndoC-βH3 cells carrying an indel in HNF1A exon 1 showed decreased HASTER as well as HNF4A—another HNF1A-dependent gene (n = 3 lentiviral transductions). The data represent means ± s.d. and are normalized to TBP. Statistical significance was determined by two-tailed Student’s t-test. j, HNF1A binds the Haster promoter in mouse liver (representative example from three replicates, MACS2 P values). The locations of seven HNF1A motifs with a JASPAR CORE score of >0.8 are shown, along with the sequences of three motifs. See Extended Data Fig. 1 for information on transcriptional start sites. k, Schematic of the HNF1A/HASTER negative feedback loop.
To examine this function in vivo, we generated mice with LoxP sites flanking a 1.8-kilobase (kb) region containing Haster transcriptional start sites (Fig. 2c and Extended Data Fig. 3a,b) and used a liver Cre transgene33 to breed liver-specific Haster homozygous deletions (HasterLKO). HasterLKO mice were born at Mendelian rates and showed normal organ formation, weight and glucose homoeostasis (Extended Data Fig. 3c,d). Consistent with human mutant cells, HasterLKO mice showed increased liver Hnf1a mRNA (1.5 ± 0.3-fold) and protein (4.5 ± 0.6-fold) (Fig. 2d–f). Similar results were observed in germline Haster mutant mice (Extended Data Fig. 3e). Thus, HASTER negatively regulates HNF1A in mouse and human hepatic cells.
HNF1A is a positive regulator of HASTER
The observation that HASTER modulates HNF1A hinted at a feedback mechanism. To examine whether HNF1A in turn regulates HASTER, we studied HNF1A-deficient cells. HASTER was strongly downregulated in pancreatic islets and liver from homozygous Hnf1a null mutant mice and in HNF1A-deficient EndoC-βH3 human β cells (Fig. 2g–i). HASTER transcripts seemed highly sensitive to HNF1A levels because partial HNF1A knockdown caused markedly decreased HASTER and only marginal changes in other HNF1A-dependent genes such as HNF4A34 (Extended Data Fig. 4a). Conversely, upregulation of Hnf1a mRNA by ~30–80% through CRISPR–Cas9 synergistic activation mediator (CRISPR–SAM) led to ~50–120% increased Haster RNA (Extended Data Fig. 4b). This effect was probably direct because the HASTER promoter has seven HNF1A recognition sequences that are bound by HNF1A in mouse liver and human EndoC-βH3 β cells (Fig. 2j). These results suggested that the HASTER promoter functions as a HNF1A-sensing platform that drives HASTER transcription in accordance with HNF1A concentrations. Taken together, our observations revealed a negative feedback loop in which HNF1A positively regulates HASTER while HASTER negatively regulates HNF1A (Fig. 2k).
HASTER negative feedback controls HNF1A pioneer-like activity
To investigate the consequences of disrupting this feedback loop, we performed RNA-seq on liver from HasterLKO and control mice (Fig. 3a and Supplementary Table 1). Consistent with the increased HNF1A levels in HasterLKO liver, deregulated transcripts and functional annotations were negatively correlated with those of Hnf1a knockout liver22 (Fig. 3b,c and Extended Data Fig. 3f). A subset of genes that were most strongly upregulated in HasterLKO liver were, however, specifically expressed in kidney or intestine—two other HNF1A-expressing organs (Fig. 3c and Extended Data Fig. 5a). Therefore, Haster mutations led to increased expression of HNF1A-dependent liver genes, but also activated ectopic transcription.
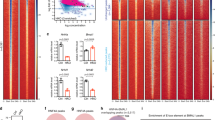
a, RNA-seq in HasterLKO liver. Differentially expressed genes (adjusted P ≤ 0.05) are highlighted in red and total numbers are indicated (n = 5 mice per genotype). FC, fold change. b, GSEA showing that genes up- or downregulated in Hnf1a KO liver have opposite expression patterns in HasterLKO liver. NES, normalized enrichment score. c, Enrichment of HasterLKO liver upregulated genes in different mouse tissues (Mouse Gene Atlas). The bars indicate Enrichr scores and the red dots show Fisher’s exact −log10-adjusted P values. d, HNF1A binding strength (log2[ChIP-seq normalized read count]) in HasterLKO and control liver (n = 3 mice). Red represents differentially bound sites (FDR ≤ 0.05) whereas blue represents a kernel density of HNF1A-bound sites with FDR > 0.05. The asterisk denotes the HasterLKO deletion. e, Left, HNF1A occupancy in control and HasterLKO liver. Right, chromatin accessibility for the same regions in liver and kidney. Neo-binding sites are bound by HNF1A only in HasterLKO. Increased bound sites include all of the other sites showing increased binding in HasterLKO. The heatmaps show the average signal of three replicates for ChIP-seq and two replicates for the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq). Windows were defined as peak centres ± 1 kb. P values were obtained with MACS2. f, Activation of a kidney-specific gene in HasterLKO liver. y axes represent MACS2 P values for ChIP-seq and RPKM for RNA-seq. g, H3K4me3 in HNF1A-bound regions in HasterLKO and control samples (average of three mice). h, Top HOMER de novo motifs for the different categories of HNF1A peak. i, Model showing that Haster KO leads to increased HNF1A (blue), causing increased HNF1A binding and expression of HNF1A-bound genes (bottom left), as well as HNF1A neo-binding sites that lead to transformation of silent inaccessible chromatin into active promoters (bottom right).
Next, we examined HNF1A genomic binding in HasterLKO liver. Overall, the HNF1A binding strength was increased in HasterLKO liver; 325 peaks showed increased HNF1A binding at a false discovery rate (FDR) of ≤0.05 (Fig. 3d). Remarkably, HasterLKO liver showed HNF1A neo-binding sites at 105 regions that were not bound by HNF1A in control livers (Fig. 3d–f).
HNF1A can bind in vitro to nucleosomal DNA35 and has been used to activate repressed liver genes in fibroblasts and reprogram them into hepatocytes36—two properties of pioneer transcription factors37. Although pioneer transcription factors have the ability to bind inaccessible chromatin, they typically show stable binding to different genomic regions across tissues22,38, suggesting that cell-specific parameters, such as perhaps cellular transcription factor concentrations, might influence their in vivo binding selectivity and the capacity to create accessible chromatin. In keeping with this notion, HNF1A neo-binding sites did not show accessible chromatin in normal liver (Fig. 3e,f), whereas they showed classical active chromatin modifications (H3K4me3 and H3K27ac) in HasterLKO liver (Fig. 3g and Extended Data Fig. 5b–f). Interestingly, HNF1A neo-binding sites contained canonical high-affinity HNF1 binding motifs, suggesting that many could be bona fide HNF1A targets in other HNF1A-expressing tissues (Fig. 3h). Thus, increased HNF1A in HasterLKO liver resulted in the creation of new binding sites, which led to the formation of new active chromatin regions.
Increased HNF1A binding at pre-existing active gene promoters in HasterLKO liver led to increased gene expression; around one-quarter of genes in this class showed greater than twofold higher expression in HasterLKO (Extended Data Fig. 5d). HNF1A neo-binding events in newly activated promoter regions led to ectopic activation of genes that are normally not expressed in liver, such as the kidney-enriched genes Ggt and Tinag (Fig. 3f and Extended Data Fig. 5d,e). Consistently, several HNF1A neo-binding sites did not show accessible chromatin in normal liver yet showed accessible chromatin in other HNF1A-expressing tissues such as kidney (Fig. 3c,f and Extended Data Fig. 5a,e). Some newly activated promoters did not overlap with any annotated mouse transcription start site, suggesting that increased HNF1A could also activate aberrant de novo promoters (Extended Data Fig. 5f,g).
In summary, genetic disruption of the HASTER feedback loop led to increased cellular HNF1A concentrations, which caused either super-activation of pre-existing HNF1A-bound promoters or the transformation of silent inaccessible chromatin into active promoters (Fig. 3i). This indicates that the HASTER feedback is crucial to control the pioneering-like activity of HNF1A, and to fine-tune the tissue specificity of HNF1A-dependent transcriptional programs.
Haster inactivation causes diabetes
HNF1A haploinsufficiency leads to pancreatic β cell dysfunction and diabetes18. To examine Haster in pancreatic cells, we used a Pdx1-Cre transgene to excise Haster in all pancreatic epithelial lineages (HasterpKO mice). HasterpKO mice showed normal morphology and growth (Extended Data Fig. 6a), yet male mice displayed glucose intolerance with insulin deficiency by 8 weeks, as well as fasting hyperglycaemia (glycaemia = 137 ± 16 mM in HasterpKO, 87 ± 5 mM in Hasterf/f littermates and 98 ± 4 mM in Pdx1-Cre; t-test P < 0.05) (Fig. 4a,b and Extended Data Fig. 6b). Male mice with germline mutations (Haster−/−) were born at Mendelian rates and showed no overt manifestations, but also showed diabetes, glucose intolerance and hypoinsulinaemia (Fig. 4c–e and Extended Data Fig. 6c,d). Thus, inactivation of Haster in the germline or in the pancreas led to impaired insulin secretion and diabetes.
a, Intraperitoneal glucose tolerance in 8-week-old male mice (n = 8 HasterpKO, n = 12 Pdx1-Cre;Haster+/+ and n = 8 Hasterf/f). P = 0.045, 8 × 10−3, 3 × 10−4, 4 × 10−4 and 5 × 10−3 at 0, 15, 30, 60 and 120 min, respectively. b, Plasma insulin of 8-week-old male mice (n = 7 HasterpKO and n = 6 Pdx1-Cre;Haster+/+). P = 0.83, 2 × 10−3 and 3 × 10−4 at 0, 15 and 30 min, respectively. c, Intraperitoneal glucose tolerance in 8-week-old male mice (n = 9 Haster−/−, n = 12 Haster+/− and n = 13 Haster+/+). P = 0.048, 0.075, 0.011, 4 × 10−4 and 2 × 10−4 at 0, 15, 30, 60 and 120 min, respectively. d, Plasma insulin in 8-week-old male mice (n = 7 Haster−/−, n = 6 Haster+/+ and n = 6 Haster+/−). P = 0.042, 0.045 and 0.026 at 0, 15 and 30 min, respectively. e, Glucose-to-insulin ratio in 8-week-old male mice (n = 9 Haster−/−, n = 12 Haster+/− and n = 13 Haster+/+). In a–e, the data are presented as means ± s.e.m. and statistical significance was determined by two-tailed Student’s t-test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). f, Immunofluorescence for HNF1A and insulin, showing either HNF1A overexpression (solid arrowheads) or no HNF1A expression (empty arrowheads) in endocrine cells of adult HasterpKO and Haster−/− mice. Note that all acinar cells from mutant mice overexpressed HNF1A (n = 3 Haster+/+, n = 3 HasterpKO and n = 2 Haster−/−). g, Immunofluorescence for HNF1A, PDX1 (a pancreatic and duodenal marker) and glucagon in Haster−/− and control E11.5 embryos, showing low heterogeneous HNF1A in pancreatic but not gut progenitors. dp, dorsal pancreas (delineated by dashed lines in KO); du, duodenum. h, Kernel density estimation of HNF1A-regulated gene expression (average z score) showing either down- or upregulation of HNF1A-dependent genes in HasterpKO HNF1Alow and HNF1Ahigh β cell clusters. i, RNA-seq (RPKM) from the indicated hESC-derived differentiation stages. j, HNF1A mRNA in hESC-derived pancreatic progenitors carrying HASTER P1 homozygous deletions (see Fig. 2a) (n = 5 independent differentiations). The data are presented as TBP-normalized relative expression (means ± s.d.). Statistical significance was determined by two-tailed Student’s t-test. k, Immunofluorescence for HNF1A, PDX1 and NKX6-1 in hESC-derived pancreatic progenitors carrying the indicated deletions, showing downregulation of HNF1A (n = 2 per deletion). In f, g and k, the scale bars represent 50 µm.
Haster knockout leads to HNF1A induction or silencing in islet cells
HasterpKO and Haster−/− pancreas showed increased HNF1A immunoreactivity in all acinar cells and in many endocrine cells (Fig. 4f). This confirmed that Haster also acts as a negative regulator of Hnf1a in the pancreas. However, numerous other islet endocrine cells from 8- to 12-week-old HasterpKO and Haster−/− mice were completely devoid of HNF1A immunoreactivity (Fig. 4f).
To further understand Haster-dependent regulation of pancreatic HNF1A expression, we analysed mice in which Haster was deleted at different stages. At embryonic stage E11.5, most Haster−/− multipotent pancreatic progenitors showed markedly heterogeneous HNF1A expression, with many cells showing low or no HNF1A expression, whereas HNF1A expression was uniform in surrounding primitive gut cells (Fig. 4g). At embryonic stage E15.5, β cells from Haster−/− and HasterpKO embryos also showed highly variable HNF1A levels, ranging from an apparent absence in many cells to marked overexpression in 1–5% of β cells (Extended Data Fig. 6e–h). This contrasted with highly uniform HNF1A staining in control embryonic β cells (Extended Data Fig. 6e,f). This dual phenotype became more evident if HasterpKO and Haster−/− mice were analysed postnatally, with more visible HNF1A-negative cells (62 and 80%, respectively) and more HNF1A-overexpressing cells (24 and 10%, respectively) (Fig. 4e,f). Inactivation of Haster after the formation of β cells, however, resulted in very few HNF1A-negative β cells and more frequent HNF1A overexpression (Extended Data Fig. 6i–k). Extended Data Fig. 6e summarizes the results from different models. Thus, Haster inactivation caused a unique variegated HNF1A expression phenotype in β cells, with co-existing silencing and overexpression. Therefore, Haster acts as a negative regulator of HNF1A in the pancreas, as in the liver, but also has a developmental cell-specific role to ensure HNF1A expression in early pancreatic progenitors and islet endocrine cells. Importantly, Haster is essential for β cell function and glucose homoeostasis.
Variegation of Haster-deficient islet cell transcriptomes
Next, we defined the transcriptional impact of HNF1A expression heterogeneity. We performed single-cell RNA-seq of islet cells from HasterpKO and control mice (Supplementary Table 2) and used graph-based clustering to separate major endocrine cell types (Extended Data Fig. 7a–c). For each cell, we calculated the average normalized expression of known HNF1A-regulated genes. Consistent with HNF1A expression heterogeneity in HasterpKO β cells, we observed increased variability of HNF1A-regulated genes across HasterpKO β cells (interquartile range = 0.53 versus 0.34 for HasterpKO and control β cells, respectively; Brown–Forsythe; P < 10−93) (Fig. 4h and Extended Data Fig. 7d–h). Further examination revealed that a large fraction of HasterpKO β cells showed increased expression of HNF1A-regulated genes, while another β cell cluster (β HNF1Alow) showed strong downregulation of HNF1A-dependent genes, such as Ttr, Tmem27, Slc2a2 and Kif12 (Fig. 4h and Extended Data Fig. 8 and Supplementary Tables 3 and 4). This β HNF1Alow cluster was specific to HasterpKO islet cells, constituted 5–21% of β cells and was discernible with independent clustering methods (Extended Data Fig. 7d–f). β HNF1Alow cells were less abundant than expected from immunostainings, possibly due to a known propensity of Hnf1a knockout cells to dissociate during islet isolation. Thus, Haster mutations caused either functional HNF1A deficiency in pancreatic β cells, which is known to cause diabetes, or overexpression of HNF1A-dependent genes. Haster, therefore, acts to ensure the stability of β cell HNF1A-regulated programs.
HASTER modulates HNF1A in human pancreatic progenitors
Next, we investigated whether HASTER also regulates HNF1A in human pancreatic cells. Analysis of published datasets showed that HASTER is activated during the early stages of hESC-derived pancreatic differentiation39 (Fig. 4i). To test HASTER function in human pancreatic progenitors, we used the hESC clones carrying the 320-bp HASTER P1 deletion (Fig. 2a) and generated pancreatic progenitors40. In contrast with the results after hepatic differentiation, which showed increased HNF1A mRNA, HASTER knockout pancreatic progenitors showed a 62% decrease of HNF1A mRNA and low heterogenous HNF1A protein levels (Fig. 4j,k). These results showed that HASTER also acts as an essential organ-specific positive regulator of HNF1A in human early pancreatic multipotent progenitor cells.
The HASTER promoter activates HNF1A in cis
Next, we explored how HASTER exerts positive and negative regulation of HNF1A, first focusing on the positive regulatory function. To assess whether HASTER acts in cis or trans, we bred compound heterozygous Hnf1a+/−;Haster+/− mice. Single heterozygous Haster+/− or Hnf1a+/− mice do not develop hyperglycaemia21 (in contrast with human HNF1A heterozygous mutations, which cause diabetes) (Fig. 5a). Remarkably, compound heterozygous Hnf1a+/−;Haster+/− young mice developed severe fasting and fed hyperglycaemia with hypoinsulinaemia, but otherwise did not exhibit extra-pancreatic manifestations observed in homozygous Hnf1a-mutant mice24,26 (Fig. 5a). This was accompanied by absent HNF1A expression in most β cells of 10-week-old Hnf1a+/−;Haster+/− mice (Fig. 5b). Because the wild-type Haster allele was not able to activate the wild-type Hnf1a, which was located on the alternative chromosome, this shows that Haster positively regulates Hnf1a in cis in islet cells. We also created hybrid-strain mice with a heterozygous Haster null allele and found decreased islet Hnf1a mRNA from the chromosome carrying the Haster null allele (P < 0.02) (Fig. 5c). Genetic experiments thus showed that Haster acts in cis to maintain Hnf1a expression in islet β cells.
a, Severe fasting and fed hyperglycaemia (left; n = 12 wild-type (WT) mice, n = 10 Haster+/− mice, n = 11 Hnf1a+/− mice and n = 13 Hnf1a+/−;Haster+/− mice) and reduced insulin secretion (right; n = 5 mice per genotype) in Hnf1a+/−;Haster+/− compound heterozygotes. The data are presented as means ± s.d. Statistical significance was determined by two-tailed Student’s t-test. b, Immunofluorescence showing normal HNF1A in Hnf1a+/− islets and no expression in most islet cells from adult Hnf1a+/−;Haster+/− mice (n = 1 per genotype). Solid arrowhead: HNF1Ahigh acinar cell. Hollow arrowhead: HNF1Alow β cell. Scale bar, 50 µm. c, Allele-specific Hnf1a mRNA in islets from hybrid-strain mice carrying the Haster mutation in the C57BL/6 chromosome. Hnf1a was quantified by strain-specific qPCR and normalized to Tbp (n = 4 mice per genotype). The data are presented as means ± s.d. Statistical significance was determined by two-tailed Student‘s t-test, d, Strain-specific RNA-seq analysis from Haster+/stop and Haster+/+ PWK/PhJ;C57BL/6 hybrid islets (n = 4 mice per genotype). RPM, reads per million reads. e, HNF1A overexpression in liver from Hnf1a+/−;Haster+/− mice (n = 1 per genotype). Scale bar, 50 µm. f, Allele-specific Hnf1a mRNA in liver from Haster+/− hybrid-strain mice carrying the Haster mutation in the C57BL/6 chromosome. Hnf1a was quantified with strain-specific assays and normalized to Tbp (n = 4 mice per genotype). The data are presented as means ± s.d. Statistical significance was determined by two-tailed Student’s t-test. g, Strain-specific RNA expression from Haster+/stop C57BL/6;PWK/PhJ hybrid mice, showing that reducing Haster elongation in liver failed to increase Hnf1a expression from the same C57BL/6 allele. The graphs show reads per million (RPM) (means ± s.d.). h, Targeting dCAS9 to the HASTER transcriptional start site blocked HASTER transcription in EndoC-βH3 cells but did not affect HNF1A or HNF4A mRNAs (n = 3 lentiviral transductions). i, CRISPR–SAM HASTER activation in EndoC-βH3 cells did not affect HNF1A and HNF4A (n = 3 lentiviral transductions). In h and i, the data represent normalized expression levels (means ± s.d.) and statistical significance was determined by two-tailed Student’s t-test.
Next, we examined whether HASTER transcriptional elongation, its RNA products or the promoter DNA are required to prevent HNF1A silencing. To this end, we created an allele with a transcriptional termination signal downstream of Haster (Hasterstop; Fig. 5d). We bred this Hasterstop allele in a hybrid-strain background and performed RNA-seq for strain-specific quantitation of Hnf1a mRNA in islets. As expected, we found severely diminished Haster transcripts from the Hasterstop allele (93% reduction; Wilcoxon rank-sum; P = 0.02). However, we still detected abundant Hnf1a exon 1 transcripts from the stop allele (Fig. 5d). Thus, whereas deletion of the Haster promoter DNA caused islet cell Hnf1a silencing in cis, this was not recapitulated by blocking Haster transcription. This indicates that the Haster promoter, but not transcriptional elongation or RNAs, is an essential positive cis-acting element of Hnf1a in pancreatic islets.
HASTER inhibits HNF1A in cis
Next, we examined how HASTER exerts negative regulation of HNF1A. To assess whether this function also occurs in cis, we again examined Hnf1a+/−;Haster+/− mice, but this time focused on liver, where Haster deficiency causes uniformly increased HNF1A expression. Compound heterozygotes showed increased HNF1A in hepatocytes, indicating that increased expression of the Hnf1a+ allele from the chromosome carrying the Haster deletion could not be compensated in trans by the Hnf1a−;Haster+ allele (Fig. 5e). Interestingly, pancreatic acinar cells showed similar behaviour to hepatocytes in compound heterozygotes, with increased HNF1A expression (Fig. 5b). We also examined Haster+/− mice bred on a hybrid-strain background and found that liver Hnf1a mRNA was selectively increased in Haster mutant chromosomes (Fig. 5f). Both findings showed that Haster-dependent inhibition of HNF1A, like its activating function, occurs in cis.
The HASTER promoter, but not its RNA, is essential for HNF1A inhibition
Next, we examined the role of HASTER transcriptional elongation, RNA molecules or its promoter in this cis-inhibitory function. Hybrid-strain mice heterozygous for Hasterstop showed that transcriptional blockage did not cause increased liver Hnf1a exon 1 transcripts in chromosomes carrying the stop allele (Fig. 5g). To further examine the role of the HASTER promoter versus transcripts, we generated clonal EndoC-βH3 cell lines with homozygous HASTER promoter deletions encompassing both transcriptional start sites (HASTERΔP/ΔP) or a 320-bp deletion of the P1 promoter (HASTERΔP1/ΔP1) (Extended Data Fig. 9a,b). Both deletions caused increased HNF1A mRNA (Extended Data Fig. 9a,b), recapitulating the phenotype of mice in which Haster was excised after the formation of β cells (Extended Data Fig. 6k). To study the role of HASTER transcription, we targeted deactivated Cas9 to the HASTER transcriptional start site (CRISPR interference (CRISPRi) roadblock41) or to a control intronic region located between HASTER and HNF1A promoters (Fig. 5h). Expectedly, targeting the HASTER promoter suppressed the formation of HASTER RNAs, although it did not influence HNF1A mRNA or HNF4A, an HNF1A-dependent transcript34 (Fig. 5h). Similarly, degradation of HASTER nuclear transcripts using GapmeRs did not affect HNF1A or HNF4A mRNAs (Extended Data Fig. 9c). Conversely, CRISPR–dCas9–SAM activation of HASTER transcription in mouse or human β cell lines led to greater than fivefold levels of HASTER RNA without changing HNF1A or HNF4A mRNAs (Fig. 5i and Extended Data Fig. 9d). Thus, modulation of HASTER transcripts or transcriptional elongation did not recapitulate the inhibitory effects of HASTER on HNF1A.
HASTER inhibition of HNF1A requires HNF1A binding to HASTER
The observation that HASTER transcriptional activation was not essential was unexpected because our genetic findings showed a tight correlation between HNF1A-dependent HASTER transcription and negative regulation of HNF1A. To reconcile these findings, we activated HASTER through lentiviral doxycycline-inducible overexpression of HNF1A (Fig. 6a). As in the CRISPR–dCas9–SAM experiments, this led to increased HASTER, but this time we observed a tenfold decrease of endogenous HNF1A mRNA (Fig. 6a). Importantly, the inhibitory effects of HNF1A overexpression were almost completely suppressed after deletion of the HASTER promoter region (Fig. 6b). Therefore, these studies showed that inhibition of HNF1A was triggered selectively by HNF1A interactions with HASTER promoter DNA, but not by various other manoeuvres that influenced HASTER transcription.
a, Doxycycline (Dox)-induced HNF1A overexpression in EndoC-βH3 cells activated HASTER and blocked endogenous HNF1A (n = 3 independent experiments). b, HNF1A overexpression in clonal EndoC-βH3 cell lines with homozygous deletions of both HASTER promoters (n = 4 independent experiments). c, HNF1A transactivation of HASTER is separable from repression of its promoter. Wild-type HNF1A or HNF1A containing a deletion of an endogenous IDR, HNF1B or HNF1B fused to an unrelated IDR were expressed in EndoC-βH3 cells. Green fluorescent protein (GFP) and GFP fused to the unrelated IDR are shown as controls (n = 4 independent experiments). NS, not significant. d, Expression of HNF1A containing a deletion of an IDR and of HNF1B fused to an unrelated IDR in EndoC-βH3 cells, essentially as represented in b with the addition of experiments with HASTER promoter deletions to show that the effects are dependent on HASTER (n = 4 independent experiments). In a–d, the data are presented as TBP-normalized relative expression (means ± s.d.) and statistical significance was determined by two-tailed Student‘s t-test.
Uncoupling of HNF1A negative autoregulation and transactivation
To further establish whether HNF1A-dependent inhibition of its own promoter was dependent on its ability to activate HASTER transcription, we selectively modified the transactivation function of HNF1A. To this end, we examined the sequence of the transcriptional activation domain of HNF1A and identified an intrinsically disordered region (IDR); IDRs have been implicated in transcriptional activation through phase separation42. A selective deletion of this IDR led to decreased HNF1A-dependent HASTER transcription, but did not prevent inhibition of HNF1A (Fig. 6c,d). We also examined HNF1B, a paralogue with the same sequence recognition specificity. We found that while HNF1B is a weaker inhibitor of HNF1A than HNF1A itself, fusion of HNF1B to an unrelated IDR from the FUS protein increased HASTER activation, yet did not have a significant impact on HNF1B-dependent HNF1A inhibition (Fig. 6c,d). Therefore, the HASTER promoter is required for HNF1A-dependent transactivation of HASTER, as well as for HNF1A autoregulation, but these are two separable molecular mechanisms.
HASTER restrains HNF1A enhancer spatial interactions
Next, we examined whether HASTER function entails changes in the local histone modification landscape. Chromatin from control liver expectedly showed localized H3K4me3 enrichment surrounding Hnf1a and Haster promoters. In contrast, HasterLKO H3K4me3 showed spreading from the Hnf1a promoter to an intronic E enhancer region (Fig. 7a). H3K4me3 was therefore significantly increased in this E region, as well as in an upstream CTCF-bound (C) region (t-test; P < 0.05) (Fig. 7b). This spreading of H3K4me3 in HasterLKO suggested that Haster might insulate the Hnf1a promoter from the intronic E enhancer, while an increase in H3K4me3 at the E and C regions in HasterLKO suggested that Haster might influence the proximity of E and C regions with the H3K4me3-rich Hnf1a promoter. We therefore hypothesized that the HASTER promoter could inhibit HNF1A by modulating three-dimensional (3D) chromatin contacts of HNF1A with local regulatory elements.
a, HasterLKO liver shows increased contacts between Hnf1a upstream viewpoints and the intronic E enhancer. UMI-4C contact trends with binomial standard deviation for the V1 and V2 viewpoints are shown (n = 6 for the wild type and n = 3 for mutant livers). Triangles denote viewpoints (DpnII fragment ± 1 kb) and asterisks mark E. The bottom panel shows liver H3K4me3. The brown shading shows the region deleted in HasterLKO. b, UMI normalized counts at E showed increased contacts with upstream regions (V1 and V2) in HasterLKO liver. Statistical significance was determined by 𝜒2 tests for n = 6 wild-type and mutant livers (V1) and n = 3 wild-type and mutant livers (V2). c, HasterLKO cells have increased H3K4me3 in C and E (n = 3 biological replicates). The data are presented as means ± s.d. Statistical significance was determined by two-tailed t-test. d, Schematic depicting increased Hnf1a promoter–E interactions in HasterLKO liver. e,f, Doxycycline-induced HNF1A overexpression in HASTER-deleted EndoC-βH3 cells (n = 4) showing (e) normalized HNF1A mRNA levels and (f) HNF1A promoter viewpoint (triangle) UMI-4C contacts. The green shading shows a 5-kb region centred on E that was used to quantify HNF1A promoter interactions. Normalized UMI counts and 𝜒2 test P values calculated with umi4c are shown on the right. g, E deletions prevent HNF1A increases in HASTER-deleted cells. HASTER+/+ or HASTERΔP1/ΔP1 clones were used to create polyclonal cells containing a mix of homozygous and heterozygous E deletions (ΔE) or wild-type sgGFP controls (WT). HASTER and HNF1A RNAs are shown as the fold change relative to parental HASTER+/+ or HASTERΔP1/ΔP1 cells. ΔE significantly reduced HASTER but not HNF1A in HASTER+/+ cells, yet it reduced HNF1A in HASTERΔP1/ΔP1 cells. Identical results were observed with a different clone, whereas C mutations had no effect (Extended Data Fig. 10f) (pool of n = 3 independent experiments with three pairs of sgRNAs for each deletion). In e and g, the data are presented as TBP-normalized relative expression (means ± s.d.) and statistical significance was determined by two-tailed t-test. h, HASTER exerts negative and positive feedbacks. At low HNF1A concentrations, HNF1A promoter–E interactions and transcription are unhindered, whereas at high HNF1A concentrations, HNF1A binding to HASTER limits HNF1A–E contacts, thereby decreasing HNF1A transcription. HASTER also acts as an essential enhancer in pancreatic lineages. i, HASTER is distinct from classic enhancers or silencers and is instead a cis-acting stabilizer that prevents overexpression and silencing.
To test this, we performed quantitative chromosome conformation capture using unique molecular identifiers (UMI-4C)43. Mouse Hnf1a and Haster promoters, as well as the intronic E enhancer region, are all located within ~7 kb. To increase the ability to capture 3D chromatin interactions with the Hnf1a 5′ region, we selected one viewpoint ~6 kb upstream of Hnf1a, near the CTCF-bound C site (viewpoint 1) and another at the Hnf1a promoter (viewpoint 2) (Fig. 7a). UMI-4C experiments from HasterLKO versus control liver (n = 6 per genotype) showed that the Haster deletion caused greater than twofold increased contacts between both Hnf1a upstream regions and the intronic E enhancer (V1; χ2 test for pooled UMI-4C libraries; P = 0.02) (Fig. 7a,c and Extended Data Fig. 10a). Thus, the analysis of two viewpoints showed consistent changes in interactions between the Hnf1a upstream region and the intronic E enhancer in HasterLKO (Fig. 7d).
Likewise, we examined human EndoC-βH3 cells that had an intact or deleted HASTER promoter region and used the HNF1A promoter as a viewpoint for quantitative UMI-4C analysis. We found that HASTER deletions caused increased interactions between the HNF1A promoter and E regions (χ2 test; P = 0.04; pooled UMI-4C libraries from four experiments). Next, we asked whether HNF1A binding to HASTER can modulate such interactions. HNF1A overexpression using the doxycycline-inducible system expectedly decreased endogenous HNF1A mRNA and significantly decreased interactions between the HNF1A promoter and the E region in HASTER+/+ cells (χ2 test; P = 0.05) (Fig. 7e,f and Extended Data Fig. 10b–d). This effect required an intact HASTER promoter, as no significant HNF1A-dependent 3D contact differences were observed in HASTER mutants (χ2 test; P = 0.78) (Fig. 7f and Extended Data Fig. 10b–d). Out of 33 enhancer-like regions in 1 megabase surrounding HNF1A, only E showed significant HNF1A-dependent changes (Extended Data Fig. 10b). Therefore, these results indicate that HNF1A overexpression limits 3D contacts between HNF1A and an intronic enhancer region, and this effect requires the HASTER promoter.
These findings imply that HASTER inhibition of HNF1A transcription involves modulation of interactions between HNF1A and the intronic E enhancer. Consistently, E deletions prevented increased HNF1A mRNA after deleting HASTER, but did not cause significant changes when HASTER was intact (Fig. 7g and Extended Data Fig. 10e,f). Taken together, these experiments show that HASTER-dependent negative feedback of HNF1A occurs through a cis function of the HASTER promoter that does not require HASTER transcription. Instead, HNF1A binding to HASTER modifies the local 3D chromatin landscape and insulates HNF1A from cis-acting intronic regulatory elements (Fig. 7h).
Discussion
These studies have uncovered a cis-regulatory element that senses HNF1A concentrations and feeds back on HNF1A to ensure appropriate cell-specific expression levels (Fig. 7h). This is achieved through a dual activating and inhibitory function that is fundamentally different from conventional cis-acting enhancers or silencers that provide spatiotemporal ON or OFF switches, respectively (Fig. 7i).
We show that HASTER’s dual function emanates from a 320-bp promoter DNA sequence and does not require transcription. However, it remains possible that transcripts have additional effects that were not explored. HASTER’s inhibitory function was triggered by high concentrations of HNF1A, which modified HNF1A promoter–enhancer interactions (Fig. 7h). The activating function of HASTER is reminiscent of an intronic enhancer, because it activates transcription in cis, and has lineage-specific essential role in pancreatic endocrine cells, plausibly due to cis-regulatory redundancy in other cell types. This dual HASTER function was most compellingly illustrated by the pancreatic knockout phenotype, in which lack of Haster enhancer-like activity led to HNF1A silencing in some β cells, while lack of negative feedback caused overexpression in other β cells that succeeded in activating HNF1A.
HASTER-dependent feedback was critical to ensure that HNF1A selects appropriate binding sites in different cell types. Interestingly, a few lncRNAs have recently been shown to negatively regulate nearby genes through different mechanisms, including the heart transcription factor gene Hand2 (refs. 12,44), the c-MYC oncogene12 or CHD2 (ref. 15). All such genes—HAND2, MYC and CHD2, as well as HNF1A—share in common that they are haploinsufficient and encode transcriptional regulators15,18,45,46. Furthermore, c-MYC, HAND2 and HNF1A have been used in misexpression systems for lineage reprogramming—a feature of transcription factors that can act on repressed chromatin36,47. These examples, and perhaps most clearly HASTER’s dual function, suggest that the principal function of a group of cis-acting lncRNA units may be to stabilize dosage-sensitive genes that encode proteins that have a capacity to transform cell-specific chromatin landscapes.
Our studies exemplify a genetic defect in a mammalian lncRNA promoter that causes an in vivo physiological phenotype. Remarkably, the main manifestation of homozygous germline Haster mutations was β cell dysfunction and diabetes. HNF1A heterozygous mutations also cause selective β cell dysfunction and only subclinical alterations in other cell types18, but homozygous Hnf1a mutations cause severe liver and renal dysfunction, growth retardation, diabetes and embryonic lethality21,24. The discovery of a transcriptional stabilizer of HNF1A that has a selective function in β cells therefore provides a lead to dissect cell-specific genetic mechanisms underlying HNF1A haploinsufficient diabetes. It is also relevant for efforts to modulate HNF1A function in β cells.
Finally, this finding has general implications for our understanding of non-coding genome defects in disease. Unlike transcriptional enhancers, which often form clusters that provide robustness to genetic disruption48,49, our findings indicate that the 320-bp HASTER promoter region lacks functional cis-regulatory redundancy. This warrants a need to examine lncRNA promoter sequence variation in human genomes.
Methods
Animal studies
Animal experimentation was carried out in compliance with EU Directive 86/609/EEC and Recommendation 2007/526/EC and enacted under Spanish law 1201/2005. Experiments were approved by the animal care committees of the University of Barcelona and Parc de Recerca Biomedica de Barcelona. Haster f (LoxP) and stop alleles were generated in C57Bl/6N JM8.F6 embryonic stem cells by homologous recombination. Briefly, mouse embryonic stem cells were electroporated with a linearized targeting plasmid containing a LoxP-flanking Haster promoter or a transcription termination (3× SV40 polyA) signal downstream of the Haster promoter, as well as a phosphoglycerate kinase/neomycin selection cassette flanked by FRT recombination sites (Extended Data Fig. 3a). Constructs were linearized by PmeI and SacII (conditional allele) or PacI and PmeI (stop allele). Electroporated embryonic stem cells were selected for the cassette with geneticin. Clones were analysed by Southern blot and targeted clones were injected into C57BL/6BrdCrHsd-Tyrc morulae (E2.5) to create chimeric mice that transmitted the recombined allele through the germline. The phosphoglycerate kinase/neomycin cassette was excised by crossings with Tg(CAG-Flp) mice. Mice were bred on C57BL/6 backgrounds unless otherwise specified.
To excise Haster in pancreatic epithelial cells, Haster+/f mice were crossed with Pdx1-Cre mice (Tg(Pdx1-Cre)6Tuv)50. Constitutive excision in β cells was achieved with Ins1Cre knock-in mice (Ins1tm1.1(cre)Thor)51. Inducible excision in β cells was achieved with the Pdx1-CreER transgene (Tg(Pdx1-Cre/Esr1*)1Mga)52 after 40 µg oral tamoxifen (Merck) twice, spaced by 4 d in 10- to 13-week-old mice, and analysed 12 weeks later. Early liver deletion was achieved with Alb-Cre mice, in which Cre is driven by an albumin promoter and alpha-fetoprotein enhancer (Alb Tg(Alb1-cre)1Khk)33. Haster germline deletions were generated by breeding Haster+/f mice with Tg(EIIa-cre)53. Hnf1a+/− mice have been described24. Genotyping primers are provided in Supplementary Table 5.
Lines with LoxP alleles without Cre, Cre lines without LoxP alleles and wild-type littermates served as controls, as indicated. Experimental cohorts were maintained on a 12 h light/12 h dark cycle with free access to water and standard mouse chow. Before decapitation, mice were anaesthetized using isoflurane (Zoetis).
Glucose tolerance
Animals were fasted overnight and received intraperitoneal glucose injections (2 g kg−1) or were re-fed before blood glucose was collected at the indicated time points. Glucose was measured with a GlucoMen Aero 2K meter (Menarini Diagnostics). Plasma insulin was quantified with the Ultra Sensitive Mouse Insulin ELISA kit (Crystal Chem) using an Infinite M Plex (Tecan) plate reader. Standard curves were fitted using quadratic polynomial regression. Assays were performed in duplicate using 5 µl plasma from mouse tail, and mean values are reported.
Islet isolation
Islet isolation was performed as described54. Briefly, ice-cold collagenase P solution (1 mg ml−1 in Hanks’ balanced salt solution (HBSS) buffer; Roche) was injected through the main duct. The inflated pancreas was dissected, incubated at 37 °C for 8 min with agitation, disaggregated by gentle suction through a needle, washed four times with cold HBSS with 0.5% bovine serum albumin (BSA) and resuspended in 7 ml 7:3 pre-cooled Histopaque 1077:Histopaque 1119 (Merck), then 7 ml HBSS with 0.5% BSA was layered on top. The gradient was centrifuged at 950g for 20 min at room temperature. The interphase containing islets was collected, washed three times with HBSS with 0.5% BSA and the islets were further enriched by aspiration under a stereomicroscope. Islets were cultured for 2 d in 11 mM glucose RPMI with 10% foetal calf serum and penicillin–streptomycin (1:100; Invitrogen) at 37 °C and under 5% CO2.
Vectors
We generated lentiviral vectors with a human insulin promoter driving the expression of Cas9 or dCas9, in addition to a U6-driven single guide RNA (sgRNA) (pLV-hIP-Cas9-BSD (plasmid 183230; Addgene) and pLV-hIP-dCas9-BSD (plasmid 183231; Addgene)). The EF1a promoter and puromycin resistance of lentiCRISPRv2 vector (plasmid 52961; Addgene) were replaced by a human insulin promoter and blasticidin-S deaminase (BSD) using Gibson Assembly (NEBuilder HiFi DNA Assembly Master Mix) to generate pLV-hIP-Cas9-BSD. The insulin promoter (343 bp) was amplified from EndoC-βH3 DNA and BSD from Addgene 61425. Cas9 from pLV-hIP-Cas9-BSD was replaced by dCas9 using Gibson Assembly to generate pLV-hIP-dCas9-BSD. dCas9 was amplified from pSp-dCas9-2A-GFP8.
sgRNAs (20 nucleotides) for CRISPRi roadblock were designed within 100 bp downstream of the islet CAGE transcriptional start site using Cas-Designer (http://www.rgenome.net/cas-designer/) and cloned as described55. Briefly, oligonucleotides (Thermo Fisher Scientific) containing sgRNAs flanked by compatible overhangs were phosphorylated with T7 polynucleotide kinase (NEB) and annealed. Oligonucleotide duplexes were ligated into BbsI- or BsmBI-digested destination vectors. Ligated constructs were transformed into Stbl3 chemically competent Escherichia coli and clones were sequenced. For deletions, sgRNA pairs were cloned as described56,57. Briefly, a fragment containing the scaffold of sgRNA1 and the H1 promoter of sgRNA2 were amplified from the pScaffold-H1 donor (118152; Addgene) with primers containing the protospacer of the sgRNA1, sgRNA2 and BbsI restriction sites. The PCR fragment was digested with BbsI and ligated into the destination vector.
A TetOn-HNF1A lentiviral vector (pLenti-CMVtight-HNF1A-FLAG-Hygro; 183232; Addgene) was built by cloning human HNF1A-FLAG into pLenti CMVtight Hygro DEST (26433; Addgene). Reverse tetracycline-controlled transactivator (rtTA) was expressed from pLV-rtTA-zeo (183233; Addgene), built by amplifying the UbC promoter and rtTA-Advance cassette from pHAGE-TRE-dCas9-KRAB (50917; Addgene) and cloned into a lentiviral backbone upstream from the 2A-ZeocinR cassette. Sequences are listed in Supplementary Table 6.
HNF1A IDRs were predicted using MobiDB-lite (http://old.protein.bio.unipd.it/mobidblite/) from InterProt. IDR1 comprised amino acids 283–358 and IDR2 comprised amino acids 545–573. Only IDR2 deletions showed significantly decreased transcription and are thus shown. Vectors carrying deletions or fusions were built using Gibson Assembly (NEBuilder HiFi DNA Assembly Master Mix) using pcDNA3.1 as the backbone (183234–183239; Addgene). A carboxy-terminal FLAG tag and the 3′ untranslated region from the Xenopus globin gene were added. The FUS IDR58 was codon optimized and synthetized as gBlock (IDT).
Cell culture
EndoC-βH3 cells59 were maintained on a 2 µg ml−1 fibronectin- and 1% extracellular matrix-coated plate in Dulbecco’s modified Eagle medium (DMEM) low glucose (1 g l−1), sodium pyruvate (Thermo Fisher Scientific), 2% BSA Fraction V (Roche), 1% heat-inactivated foetal bovine serum (FBS; Labtech), 2 mM l-glutamine, 5.5 μg ml−1 human transferrin, 1 mM sodium pyruvate 10 mM nicotinamide, 6.7 ng ml−1 sodium selenite, 50 μM β-mercaptoethanol, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin. DMEM was substituted with Advance DMEM/F-12 (Thermo Fisher Scientific) and FBS was omitted for the TetOn-HNF1A EndoC-βH3 cell line, as well as during the expansion of EndoC-βH3 clones.
293FT cells (Thermo Fisher Scientific) were maintained in DMEM, 10% heat-inactivated FBS, 0.1 mM MEM non-essential amino acids, 2 mM l-glutamine, 1 mM sodium pyruvate, 500 µg ml−1 geneticin, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin.
MIN6 cells60 were maintained in DMEM, 4.5 g l−1 glucose, 15% heat-inactivated FBS, 50 μM β-mercaptoethanol and 50 µg ml−1 gentamicin.
Gene perturbations in EndoC-βH3 cells
LNA GapmeRs (Exiqon; Supplementary Table 7) and plasmid vectors were nucleofected in EndoC-βH3 cells using Nucleofector B2 with an Amaxa Cell Line Nucleofection Kit V (Lonza) and program G-017. We used 2 million cells and 10 µg plasmid DNA per nucleofection for deletions or 1 million cells with 250 pg LNA GapmeRs. Cells were harvested 72 h after GapmeR or 48 h after plasmid nucleofection.
CRISPRi and CRISPR–SAM lentiviral particles were produced as described56. 293FT cells were seeded at 75,000 cells per cm2 in T75 flasks and, 24 h later, transfected with CRISPR and packaging plasmids pMDLg/pRRE, pRSV-Rev and pMD2.G (12251, 12253 and 12259; Addgene) with PEIpro (Polyplus-transfection) in antibiotic-free media using a 1:1 ratio of total µg DNA to µl PEIpro. The medium was replaced with 9 ml fresh 293FT antibiotic-free media 18 h post-transfection and lentiviral particles were collected 72 h post-transfection. Immediately after collection, the supernatants were centrifuged for 5 min at 400g and filtered using 0.45-µm pore size Steriflip-HV polyvinylidene fluoride filters (Millipore). The supernatant was supplemented with 1 mM MgCl2 and treated with 1 µg ml−1 DNase I (Roche) for 20 min at 37 °C. Viral particles were concentrated with 1:3 vol/vol of Lenti-X Concentrator (Clontech) at 4 °C overnight. On the following day, virus particles were collected for 45 min at 1,500g and 4 °C, resuspended in 100 µl phosphate-buffered saline (PBS), aliquoted and stored at −80 °C. Transduction was carried out with 10 µl virus for 400,000 cells in 1 ml. Antibiotic selection was started 3 d later with 8 µg ml−1 blasticidin, 100 µg ml−1 hygromycin or 200 µg ml−1 zeocin for EndoC-βH3 cells.
A CRISPR–SAM cell line was established by successive transduction of lentivirus dCAS-VP64_Blast (61425; Addgene) and MS2-P65-HSF1_Hygro (61426; Addgene). Cells were transduced with lentivirus sgRNA(MS2)_zeo (61427; Addgene) expressing gene promoter-targeting sgRNAs. The CRISPR–Cas9 and CRISPRi roadblock experiments were performed with lentivirus hIP-Cas9-BSD (pLV-hIP-Cas9-BSD plasmid) and hIP-dCas9-BSD (pLV-hIP-dCas9-BSD plasmid).
For the CRISPR–Cas9 clonal deletions, EndoC-βH3 cells were nucleofected with pSpCas9(BB)-T2A-HygR (118153; Addgene) containing sgRNAs. At 24 h after nucleofection, cells were selected using Hygromycin B (200 µg ml−1; Thermo Fisher Scientific; 10687010) for 3 d. After 2 weeks, the cells were seeded at low density (2–9 cells per cm2) in Advance DMEM/F-12-based EndoC-βH3 medium (Gibco). EndoC-βH3 clones were hand picked and transferred into 96-well plates. After genotyping, selected clones were expanded in DMEM-based 1% FBS medium.
Doxycycline-inducible HNF1A EndoC-βH3 cells were established by successive transduction with rtTA-2A-ZeoR-expressing lentivirus (pLV-rtTA-zeo) and TRE-HNF1A-FLAG lentivirus carrying the hygromycin resistance (pLenti-CMVtight-HNF1A-FLAG-Hygro). Cells were exposed to doxycycline (0, 25, 50, 100, 200 or 400 ng ml−1) for 24 h and endogenous HNF1A mRNA was detected by PCR with oligonucleotides recognizing a 3′ untranslated region that is not present in HNF1A-FLAG. Exogenous HNF1A-FLAG was detected by PCR with oligonucleotides specific for the FLAG region.
hESC genome editing
H9 hESCs were maintained in mTeSR1 medium (85870; STEMCELL Technologies) on a Matrigel (356231; Corning)-coated plate. For nucleofection, cells were dissociated with Accutase (Merck) for 8 min at 37 °C, diluted in 10 µM Y27632 (Merck) mTeSR1, centrifuged at 110g for 3 min and resuspended in 10 µM Y27632 mTeSR1. A total of 106 cells were nucleofected using Human Stem Cell Nucleofector Kit 2 (program G-017; Lonza) with 5 µg pSpCas9(BB)-2A-puro (62988; Addgene) expressing two sgRNAs. After nucleofection, the cells were transferred to a 12-well plate containing 1 ml 10 µM Y27632 mTeSR1. After 24 h, the cells were selected for puromycin resistance by replacing the medium with 10 µM fresh Y27632 mTeSR1 containing 0.5 µg ml−1 puromycin for 24 h. After selection, the hESCs were cultured in mTeSR1 without Y27632. After two passages, the cells were dissociated and plated at low density. Isolated clones were transferred and maintained in 96-well plates until genotyping.
hESC differentiation
H9 mutant clones were differentiated to hepatocytes using a protocol adapted from Hannan et al.32. Cells were seeded at 300,000 cells per 24-well plate in 10 µM Y27632 mTeSR1 and differentiation was started after 24 h. The following media were used for differentiation: (1) S1 medium61 was prepared with MCDB 131 Medium (10372019; Thermo Fisher Scientific) supplemented with 8 mM d-(+)-Glucose (G7528; Merck), 2.46 g l−1 NaHCO3 (S3817; Merck), 2% BSA Fraction V (10735078001; Roche), 1:50,000 Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) (51500056; Thermo Fisher Scientific), 2 mM GlutaMAX (35050061; Thermo Fisher Scientific) and 0.25 mM l-ascorbic acid (A4544; Merck); (2) RPMI/B27 medium was prepared with RPMI 1640 Medium, GlutaMAX Supplement (61870010; Thermo Fisher Scientific) supplemented with B-27 Supplement (17504044; Thermo Fisher Scientific) and MEM Non-Essential Amino Acids Solution (11140035; Thermo Fisher Scientific); and (3) hepatocyte growth medium (HGM) was prepared with HBM Basal Medium (CC-3199; Lonza) supplemented with 3.75 g ml−1 BSA Fraction V, 250 µg ml−1 l-ascorbic acid, 10 µg ml−1 holo-Transferrin (T0665; Merck), 0.5 µg ml−1 Hydrocortisone (H0888; Merck), 5 µg ml−1 human Insulin and 10 ng ml−1 epidermal growth factor (236-EG-200; R&D Systems). During differentiation, the medium was changed every day, or every 2 d after day 11, using the following media: S1 medium with 100 ng ml−1 Activin A (338-AC-050; R&D Systems) and 3 μM CHIR 99021 (04-0004; Tocris Bioscience) for day 1; S1 medium with 100 ng ml−1 Activin A for days 2 and 3; RPMI/B27 medium with 50 ng ml−1 Activin A for days 4–6; RPMI/B27 medium with 20 ng ml−1 BMP-4 (314-BP-010; R&D Systems) and 10 ng ml−1 FGF-10 (ABE1324; Source BioScience) for days 7–10; and HGM medium with 30 ng ml−1 Oncostatin M (295-OM-010; R&D Systems) and 50 ng ml−1 HGF (100-39; PeproTech) for days 11–25. The definitive endoderm stage was reached at day 4, the anterior endoderm stage was reached at day 7, the hepatoblast stage was reached at day 11 and the hepatocyte stage was reached at day 26.
Pancreatic differentiations were performed using a modification of a published protocol40. Dissociated hESCs were seeded at 2 million cells per 35 mm well coated with Matrigel in 5 µM Y27632 E8 medium (A1517001; Thermo Fisher Scientific). Differentiation was started the following day after washing the cells once with 1× PBS: Definitive endoderm induction was as follows: MCDB 131, 2 mM GlutaMax (35050038; Thermo Fisher Scientific), 1.5 g l−1 NaHCO3, 0.5% BSA Fraction V (7500804; Lampire Biological Laboratories), 10 mM final glucose, 100 ng ml−1 Activin A (QK001; Qkine) and 3 µM CHIR 99021 (4423; Tocris Bioscience) on day 0; as for day 0 but reducing CHIR 99021 to 0.3 µM on day 1; and as for day 1 but with no CHIR 99021 on day 2. Stage 2 posterior foregut induction was as follows: MCDB 131, 2 mM GlutaMax, 1.5 g l−1 NaHCO3, 0.5% BSA Fraction V, 10 mM final glucose, 0.25 mM ascorbic acid (A4544; Sigma–Aldrich) and 50 ng ml−1 FGF-7 (Z03407-1; GenScript) on days 3–5. Stage 3 pancreatic endoderm induction was as follows: MCDB 131, 2 mM GlutaMax, 2.5 g l−1 NaHCO3, 2% BSA Fraction V, 10 mM final glucose, 0.25 mM ascorbic acid, 50 ng ml−1 FGF-7, 0.25 µM SANT-1 (S4572; Sigma–Aldrich), 1 µM retinoic acid (R2625; Sigma–Aldrich), 100 nM LDN193189 (S2618; Selleckchem), 1:200 ITS-X (51500056; Thermo Fisher Scientific) and 200 nM TPB (sc-204424; Santa Cruz Biotechnology) on days 6 and 7. Stage 4 pancreatic progenitor induction was as follows: MCDB 131, 2 mM GlutaMax, 2.5 g l−1 NaHCO3, 2% BSA Fraction V, 10 mM final glucose, 1:200 ITS-X, 0.25 mM ascorbic acid, 2 ng ml−1 FGF-7, 0.25 uM SANT-1, 0.1 µM retinoic acid, 200 nM LDN, 100 nM TPB, 100 ng ml−1 epidermal growth factor (AF-100-15; PeproTech), 10 mM nicotinamide (N0636; Sigma–Aldrich), 10 ng ml−1 Activin A and 10 µM Y27632 on days 8–11. Cells were dissociated with TrypLE and seeded in AggreWell 400 plates (34425; Stem Cell Technologies) on day 10.
Reverse transcription quantitative PCR
RNA was prepared using an RNeasy Mini Kit (Qiagen) and DNAse I (Qiagen) and retrotranscribed with SuperScript III (Thermo Fisher Scientific) and random hexamers (Thermo Fisher Scientific). Quantitative PCR was performed with Universal Probe Library assays (Roche). Reactions were carried out in duplicate in a QuantStudio 12K Flex (Applied Biosystems) with 1× TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), 1 µM forward and reverse primers and 250 nM Universal Probe Library probe, or 1× TaqMan assay. Quantification was performed using standard curves, with duplicate means reported, normalized by TBP or RPLP0, as indicated. Oligonucleotides are listed in Supplementary Table 5.
Single-molecule fluorescence in situ hybridization
Single-molecule fluorescence in situ hybridization was performed as described62. A set of 48 probes (Supplementary Table 8), coupled with Quasar 570 (548/566) or Quasar 670 (647/670), were designed for each transcript (Stellaris RNA FISH probes; LGC Biosearch Technologies). EndoC-βH3 cells were grown on coated (2 µg ml−1 fibronectin and 1% extracellular matrix; Merck) coverslips. Cells were fixed in 4% formaldehyde for 2 min, washed with 1× PBS and permeabilized with 70% ethanol at 4 °C for >1 h. Probes were hybridized overnight at 37 °C in the dark with 10% formamide, 100 mg ml−1 dextran sulfate, 2× SSC and 12.5 µM probes. The following day, cells were washed for 30 min at 37 °C with 10% formamide and 2× SSC, followed by 30 min with 5 ng ml−1 4′,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted using VECTASHIELD HardSet mounting media. Acquisitions were performed on a Zeiss Axio Observer inverted widefield microscope with light-emitting diode illumination. Z-stack acquisitions were taken with a 63× objective every 0.5 μm from a total depth of 40 μm and deconvoluted (Huygens Software) and maximal projections of whole stacks were used for counting (8–12 fields per sample).
Immunofluorescence
Embryos and adult tissues were processed for immunofluorescence as described63. Briefly, tissues were fixed in 4% paraformaldehyde overnight at 4 °C, then washed in PBS before paraffin embedding. Deparaffinized sections (4 µm) were incubated for 30 min in antibody diluent (Dako) with 3% normal serum from the same species as the secondary antibody, incubated overnight at 4 °C with primary antibody and then overnight at 4 °C with secondary antibody, then DAPI stained and mounted with Mounting Medium (S3023; Molecular Probes). The primary antibodies were: HNF1A (1:400; D7Z2Q; Cell Signaling Technology), insulin (1:200; A0564; Dako), glucagon (1/1,000; 4030-01F; Millipore), Cytokeratin 19 (1/100; TROMA-III-c; Hybridoma Bank), PDX1 (1:200; AF2419; R&D Systems) and NKX6.1 (1:200; F55A10; Hybridoma Bank). The following secondary antibodies were used: Donkey anti-rabbit Alexa Fluor 488 (1/800; 711-545-152; Jackson ImmunoResearch) and Cy3 (1/400; 711-166-152; Jackson ImmunoResearch), Donkey anti-guinea pig Alexa Fluor 488 (1/800; 706-545-148; Jackson ImmunoResearch) and Cy5 (1/400; 706-175-148; Jackson ImmunoResearch), Donkey anti-goat Alexa Fluor 488 (1/800; 705-545-147; Jackson ImmunoResearch), Donkey anti-rat Cy3 (1/400; 712-165-153; Jackson ImmunoResearch) and Donkey anti-mouse Cy5 (1/400; 715-175-151; Jackson ImmunoResearch). Images were acquired using a Leica TSE confocal microscope for tissues and a Leica DMi8 for cell lines.
Western blots
Proteins were extracted from frozen mouse livers with 9 M urea. Quantification was performed using a Microplate BCA Protein Assay Kit (23250; Thermo Fisher Scientific). Western blot was performed with 20 μg protein on 4–12% Bis-Tris gel (NP0335BOX; Thermo Fisher Scientific) using β-tubulin (2146; Cell Signaling Technology) and HNF1A antibodies (89670; Cell Signaling Technology) and Goat Anti-Rabbit IgG H&L (HRP) (1/2,000; ab97051; Abcam).
Cellular fractionation
Cellular fractionation was performed as described64. Some 5 million EndoC-βH3 cells were incubated for 5 min on ice in 200 µl cold lysis buffer (10 mM Tris-HCl pH 7.5, 0.05% IGEPAL, 150 mM NaCl and 100 U ml−1 SuperaseIn (Thermo Fisher Scientific)). The lysate was layered over 2.5 volumes of chilled sucrose solution (10 mM Tris-HCl pH 7.5, 0.05% IGEPAL, 150 mM NaCl, 24% sucrose and 100 U ml−1 SuperaseIn) then centrifuged for 10 min at 15,000g and 4 °C. The cytoplasmic supernatant was kept and the pellet was washed with 500 µl wash buffer (1 mM ethylenediaminetetraacetic acid (EDTA) in PBS pH 7.5), then centrifuged for 10 min at 15,000g and 4 °C. This pellet was resuspended in 100 µl cold glycerol buffer (20 mM Tris-HCl pH 7.5, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM dithiothreitol, 1× protease inhibitor cocktail (Roche), 50% glycerol and 100 U ml−1 SuperaseIn), and 100 µl cold nuclei lysis buffer (10 mM HEPES pH 7.5, 1 mM dithiothreitol, 7.5 mM MgCl2, 0.2 mM EDTA pH 8, 0.3 mM NaCl, 1 M urea and 1% IGEPAL) was added to the nuclei suspension, vortexed and left on ice for 2 min. Nuclear lysate was centrifuged for 2 min at 15,000g and 4 °C and the supernatant was collected as nucleoplasmic fraction. The pellet (chromatin fraction) was washed with 500 µl wash buffer and resuspended in 300 μl chromatin DNase buffer (20 mM Tris-HCl pH 7.5, 50 mM KCl, 4 mM MgCl2, 0.5 mM CaCl2, 2 mM TCEP (Merck), 1× protease inhibitor cocktail, 0.4% sodium deoxycholate, 1% IGEPAL and 0.1% N-lauroylsarcosine). Next, 15 µl murine RNase inhibitor (NEB) and 30 μl TURBO DNase (Ambion) were added and the reaction was incubated for 20 min at 37 °C. DNase was inactivated with 12.5 µl 25× Stop Solution (250 mM EDTA and 125 mM ethylene glycol tetraacetic acid). Proteins were digested with 7.5 μl proteinase K (Ambion) for 1 h at 37 °C. RNA from the different fractions was purified using an RNA Clean & Concentrator-25 Kit (Zymo Research).
3′ RACE
3′ RACE was performed as described65. Human islet RNA (240 ng was retrotranscribed with QT primers using SuperScript III. Nested PCRs were performed with Q5 polymerase (NEB). The first PCR used one-twentieth of complementary DNA with a gene-specific forward primer 1 and a QO reverse primer, while the second PCR used 1 µl of a 1:5 dilution of the first PCR with a gene-specific forward primer 2 and a QI reverse primer. The resulting fragments were cloned and Sanger sequenced. Oligonucleotides are provided in Supplementary Table 5.
Chromatin immunoprecipitation
Liver was collected after perfusion of ice-cold PBS and minced with a razor blade. Minced liver (100 mg) or 100–500 mouse islets were incubated with 1% formaldehyde (Agar Scientific) for 10 min at room temperature, then one-tenth of 1.25 M glycine was added for 5 min at room temperature, pelleted at 800g and 4 °C for 3 min and washed twice with PBS. Aliquots containing 20 mg initial liver or all processed islets were snap-frozen and stored at −80 °C until use. Crosslinked samples were lysed using ice-cold 2% Triton X-100, 1% sodium dodecyl sulfate (SDS), 100 mM NaCl, 10 mM Tris-HCl pH 8, 1 mM EDTA pH 8 and 1× protease inhibitor cocktail for 15–20 min on ice. Chromatin was sonicated with a Covaris S220 Focused-ultrasonicator (2% duty factor; 105 W peak incident power; 200 cycles per bust; 16 min). Sheared chromatin was centrifuged at full speed for 10 min at 4 °C to remove debris and insoluble chromatin and the supernatant was transferred to a fresh low-binding tube. For liver, the chromatin equivalent of 5 µg DNA was used for one-histone-mark chromatin immunoprecipitation (ChIP) and 10 µg was used for transcription factor ChIP. Chromatin was diluted four times with ChIP Dilution Buffer (0.75% Triton X-100, 0.1% sodium deoxycholate, 140 mM NaCl, 50 mM HEPES pH 8, 1 mM EDTA and 1× protease inhibitor cocktail) and 5% was used as input. Dynabeads Protein G (30 µl; Thermo Fisher Scientific) were blocked with BSA overnight at 4 °C. HNF1A antibody (10 µl; D7Z2Q; Cell Signaling Technology), 2 µg H3K27ac antibody (ab4729; Abcam) and 2 µg H3K4me3 antibody (15-10C-E4; Merck) or 2 µg H3K4me1 antibody (ab8895; Abcam) were added to 500 µl samples and incubated overnight with rotation at 4 °C. Magnetic beads (30 µl) were added to the samples and rotated at 4 °C for 2 h.
For ChIP-quantitative PCR (ChIP-qPCR), antibody-incubated samples were washed with low-salt wash buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 20 mM Tris-HCl pH 8 and 2 mM EDTA pH 8), high-salt wash buffer (1% Triton X-100, 0.1% SDS, 500 mM NaCl, 20 mM Tris-HCl pH 8 and 2 mM EDTA pH 8), LiCl wash buffer (0.25 M LiCl, 1% IGEPAL, 1% sodium deoxycholate, 10 mM Tris-HCl pH 8 and 1 mM EDTA pH 8) and three times with TE buffer. Elution was performed with 200 µl 1% SDS and 0.1 M NaHCO3 for 30 min at room temperature. Samples were placed on a magnet and the supernatant was transferred to a new tube. RNase A (1 µl; Thermo Fisher Scientific) was added to the eluate and incubated for 30 min at 37 °C. Reverse crosslink was performed by adding 8 µl 5 M NaCl and 3 µl proteinase K (Thermo Fisher Scientific) and incubation was performed for 1 h at 55 °C and 1,200 r.p.m., then overnight at 65 °C and 1,200 r.p.m. DNA was purified using a MinElute PCR Purification Kit (Qiagen). Quantitative PCR was carried out in duplicates as described for reverse transcription qPCR. Allele-specific qPCR was performed using Custom TaqMan SNP Genotyping Assays. Enrichment was subsequently normalized by the input.
For ChIPmentation, washes and tagmentation were performed as reported66. Antibody-incubated samples were washed twice with RIPA-LS (10 mM Tris-HCl pH 8, 140 mM NaCl, 1 mM EDTA pH 8, 0.1% SDS, 0.1% sodium deoxycholate and 1% Triton X-100), twice with RIPA-HS (10 mM Tris-HCl pH 8, 500 mM NaCl, 1 mM EDTA pH 8, 0.1% SDS, 0.1% sodium deoxycholate and 1% Triton X-100), twice with RIPA-LiCl (10 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA pH 8, 0.5% IGEPAL and 0.5% sodium deoxycholate) and once with 10 mM Tris-HCl pH 8. Beads were resuspended in 20 µl tagmentation solution (10 mM Tris-HCl pH 8, 5 mM MgCl2 and 10% vol/vol dimethylformamide) containing 1 µl Tn5 (Illumina) and incubated at 37 °C for 10 min. The reaction was stopped with 1 ml ice-cold RIPA-LS for 5 min on ice. Beads were washed twice with RIPA-LS and twice with TE buffer and resuspended in elution buffer (10 mM Tris-HCl pH 8, 5 mM EDTA pH 8, 300 mM NaCl and 0.4% SDS). Proteinase K was added to the elution and incubated for 1 h at 55 °C and 1,200 r.p.m., then overnight at 65 °C and 1,200 r.p.m. DNA was purified using a MinElute PCR Purification Kit (Qiagen). To estimate the number of cycles required for library amplification, 2 µl of the elution was used for SYBR Green qPCR, using KAPA HiFi polymerase (Kapa Biosystems). The resulting Ct value plus 1 cycle was used as the number of cycles to amplify the library. Libraries were amplified from 20 µl of elution with KAPA HiFi polymerase and Nextera custom primers (Supplementary Table 5). DNA clean-up was performed with 1.8× volume and size selection with a 0.65× volume of AMPure XP beads (Beckman Coulter). Libraries were sequenced on a HiSeq 2500 using 1 × 50 bp reads.
ChIP sequencing
ChIP sequencing (ChIP-seq) reads were aligned with Bowtie 2 (version 2.3.5) on the GCRm38 genome and sorted using SAMtools (version 1.7). Alignment statistics are listed in Supplementary Table 2. Multi-mapped reads were discarded. Reads mapping to ENCODE blacklisted regions were removed using BEDTools (version 2.27.1) and duplicated reads were removed with Picard (version 2.6.0). Peak calling was performed using MACS2 (version 2.1.1) with an FDR (q value) threshold of 0.05. The --broad flag was used for histone modifications. The MACS2 bdgcmp function was used to generate the local Poisson test −log10[P values]. P value BedGraphs were converted to bigWig using bedGraphToBigWig. Differential binding was performed using DiffBind (version 2.8.0) on peaks called in at least two samples from any genotypes, using normalized read coverage from triplicates. Binding differences were determined at q ≤ 0.05. HNF1A neo-binding sites were defined as peaks observed in at least two Haster knockout samples (q ≤ 0.05), without significant peaks in any control sample and with average log2-normalized ChIP read counts of ≤2 in control samples. Activated promoters were similarly defined as H3K4me3 peaks (q ≤ 0.05) in two Haster knockout and no control samples, with log2 normalized counts of <2 in controls and significant differential H3K4me3 enrichment (q ≤ 0.05) in Haster versus controls. Coverage was calculated using deepTools (version 3.0.2) computeMatrix and the average of the three replicates was calculated for each bin. Peak intersections were performed with pybedtools (version 0.8.0).
Motif analysis
Analysis of known and de novo transcription factor binding site motifs was performed using HOMER (version 3.12). Analyses were performed on the merge between overlapping consensus peaks defined by DiffBind, using a minimum overlap of 1 bp. Enrichment analysis of de novo transcription factor motifs was also performed with the findMotifsGenome.pl command on consensus peaks defined by DiffBind, for lengths of 8, 10 and 12 bp on the masked mm10 genome.
Assay for transposase-accessible chromatin with high-throughput sequencing
Reads were trimmed to remove adaptors using Trim Galore and aligned with Bowtie 2 (version 2.3.5) on the GCRm38 genome. Multi-mapped and duplicated reads were removed using Picard (version 2.6.0). Mitochondrial and ENCODE blacklisted region reads were discarded. For visualization, MACS2 bdgcmp was used to generate the local Poisson test −log10[P value] bedGraphs. BedGraphs were converted to bigWig using bedGraphToBigWig. The coverage was calculated using deepTools (version 3.0.2) computeMatrix for 1-kb windows with 10-bp bins.
RNA-seq
RNA from islets or liver was quantified with Qubit (Thermo Fisher Scientific) and verified with Bioanalyzer (Agilent). Libraries were prepared with a TruSeq Stranded mRNA Library Kit and sequenced on a HiSeq 4000 (2 × 75 bp reads). Reads were aligned to the GCRm38 genome with STAR (version 2.3.0). Transcript-level quantification was performed with Salmon (version 0.11) using GENCODE GCRm38 VM18 annotations (Supplementary Table 2). Gene-level normalization and differential expression were performed using the Bioconductor R (version 3.6.1) package DESeq2 (version 1.24.0), using adjusted P ≤ 0.05 as a cut-off for differentially expressed genes. Fold changes were adjusted with lfcShrink using the apeglm option67.
For differential expression, de novo transcripts from HasterLKO and control liver were assembled from RNA-seq using StringTie (version 2.0). Transcripts from HasterLKO and control replicates were merged in a single GTF file using gffcompare (version 0.10.1). Transcript quantification and differential transcript expression were performed using Salmon and DESeq2 as described above, using the merged HasterLKO and control liver transcriptome as a reference. Transcripts with low abundance (mean normalized transcripts per million< 3) were discarded. To define transcripts with an HNF1A-bound promoter, a minimum overlap of 1 bp between the transcription start site and an HNF1A peak was required.
Human, chicken, Xenopus tropicalis and zebrafish liver RNA-seq reads were aligned on the GCRh37 (hg19), galGal5, XenTro9 and GRCz11 (danRer11) genomes, respectively. Mouse kidney and small intestine RNA-seq reads were aligned to the GCRm38 (mm10) genome.
Allele-specific RNA-seq
Stranded total RNA libraries from C57BL/6;PWK/PhJ F1 liver and islets were sequenced on a HiSeq 2500 using 2 × 125 bp reads. Reads were aligned by STAR (version 2.7.6) using WASP68, and reads that aligned to a different genomic region after swapping the C57BL/6 variant to the PWK/PhJ variant were discarded. PWK/PhJ single-nucleotide polymorphisms were obtained from the Mouse Genome Project (version 5)69. ASEReadCounter (GATK version 4.1.9.0) was used to count the reads overlapping Hnf1a and Haster PWK/PhJ exonic single-nucleotide polymorphisms.
HNF1A-regulated gene set
To define a high-confidence islet HNF1A-dependent gene set, we intersected: (1) 115 downregulated genes from Hnf1a−/− islets22; and (2) 570 genes that showed increased expression (adjusted P ≤ 0.05 and mean normalized expression > 500) after CRISPR–SAM activation of Hnf1a in MIN6 mouse β cells. This MIN6-SAM cell line was generated by successive transduction of MIN6 cells with lentivirus dCAS-VP64_Blast (61425; Addgene), followed by blasticidin selection (1 µg ml−1) and lentivirus MS2-P65-HSF1_Hygro (61426; Addgene), followed by hygromycin selection (100 µg ml−1). MIN6-SAM cells were subsequently transduced with sgRNA expressing vector lenti sgRNA-(MS2)-zeo (61427; Addgene). RNAs from triplicates of two independent Hnf1a-activating sgRNAs and two independent control sgRNAs were used for RNA-seq. In total, 21 genes showed concordant downregulation and upregulation in both models (Supplementary Table 9).
Gene set enrichment analysis and Enrichr
Gene set enrichment analysis (GSEA) was performed with GSEAPreranked (version 6.0; GenePattern)70 on genes ranked by fold change, using default parameters over 10,000. Enrichments of functional annotations were performed with Enrichr71.
Tissue-specificity z score
Tissue-specificity z scores were calculated for each gene by taking the average normalized gene expression in tissue minus the mean of all Hnf1a-expressing tissues divided by the standard deviation of all Hnf1a-expressing tissues72.
Single-cell RNA-seq
Cultured mouse islets were dissociated with Accutase (Merck) for 15 min at 37 °C. Islet cell suspensions were centrifuged at 600g for 3 min and resuspended in culture medium with DAPI before FACS sorting to remove dead cells and doublets. After sorting, cells were centrifuged at 600g for 3 min and resuspended in PBS/0.04% BSA. Single-cell libraries were generated with a 10X Genomics Chromium Single Cell 3′ Reagent Kit v3 following the manufacturer’s instructions. Libraries were sequenced on a HiSeq 4000 using 2 × 75 bp reads.
Single-cell RNA-seq analysis
Read alignments and UMI counts were performed with CellRanger (version 3.0.2) using the mm10 reference genome. Subsequent analyses were carried out with Seurat (version 3.0.1)73 or scVI-tools (version 0.11.0)74.
For Seurat, cells with <500 genes or >5% mitochondrial genes were filtered out (Supplementary Table 2). UMI counts were normalized using SCTransform75. To define shared populations between controls and knockouts, we performed an integrated analysis on the three control and three knockout datasets76. Briefly, the 3,000 most variable genes were used to find anchors (SelectIntegrationFeatures function using 50 dimensions). The first 50 principal components were used for t-distributed stochastic neighbour embedding projection (RunTSNE function) and clusters were defined by graph-based unsupervised clustering (FindClusters function) with a resolution of 0.5.
For scVI analysis, cells with <1,000 or >6,000 genes or >5% mitochondrial genes were filtered out. UMI counts were normalized for library size and log transformed. Integration of control and knockout samples was performed using the top 2,000 variable genes. The scVI model was trained using ten dimensions of latent space and two hidden layers for the encoder and decoder neural network. The identification of HNF1A-deficient β cell clusters was robust to using Seurat or scVI (Extended Data Fig. 7).
Differential expression was performed with Seurat FindMarkers (min.pct = 0.1) for all combinations of controls versus knockouts. Wilcoxon rank-sum P values from the different combinations were combined using Fisher’s method. Only genes with a consistent positive or negative fold change across all control or knockout combinations and with a combined P ≤ 0.05 were considered differentially expressed. All genes differentially expressed in endothelial cells were discarded. For differential expression, all β cell clusters with >250 cells were grouped in a single β cluster.
Seurat objects were exported as loom using as.loom of the loomR (version 0.2.0.1) library. Data visualization was performed with Python (version 3.7.3) and loompy (version 2.0.16), NumPy (version 1.15.4), pandas (version 0.25.0), Matplotlib (version 3.1.0) and seaborn (version 0.9.0) libraries. Statistics were computed using SciPy (version 1.1.0).
UMI-4C
UMI-4C was performed as described43 with modifications. Liver from three samples per genotype was crosslinked with 2% formaldehyde for 10 min, as described for ChIP. EndoC-βH3 cells were fixed with 1% formaldehyde for 10 min. Frozen pellets of ~107 cells were thawed on ice and resuspended in 5 ml cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% IGEPAL and 1× protease inhibitor cocktail). After isolation, nuclei were resuspended in 650 µl nuclease-free water, 60 µl DpnII buffer and 15 µl 10% SDS and incubated at 37 °C and 900 r.p.m. for 1 h, with an additional hour after the addition of 75 µl 20% Triton X-100. The chromatin was digested at 37 °C and 900 r.p.m. for 24 h using 600 U DpnII (R0543L; NEB) and the enzyme was inactivated by incubating at 65 °C for 20 min. Ligation was performed in a final volume of 7 ml with 60 U T4 DNA ligase (Promega) and incubated at 16 °C overnight. The efficiency of the digestion and ligation was assessed by gel electrophoresis. Chromatin was reverse crosslinked with 30 µl proteinase K (10 mg ml−1) overnight at 65 °C, followed by 45 min of incubation with 30 µl RNase A (10 mg ml−1) at 37 °C. The DNA was purified by phenol–chloroform extraction followed by ethanol precipitation and resuspended in 10 mM Tris-HCl pH 8. Then, 10 µg DNA was sonicated using an S220 Focused-ultrasonicator (Covaris) to obtain 400- to 600-bp fragments. The DNA was end-repaired with 10 µl NEBNext End Repair Mix (E6050L; NEB) in a final volume of 200 µl, incubated for 30 min at 20 °C, purified with 2.2× AMPure XP beads (Beckman Coulter) and eluted in 10 mM Tris-HCl pH 8. A-tailing was performed with 200 U Klenow Fragment (M0212M; NEB) in 100 µl 1× NEBuffer 2 with 1 nM dATP. 5′ ends were dephosphorylated at 50 °C for 60 min with 20 U calf intestinal alkaline phosphatase (M0290S; NEB). The DNA was then cleaned with 2× AMPure XP beads. Adaptors were ligated with 0.4 µM Illumina-compatible forked indexed adaptors (Supplementary Table 5) and 10 µM quick ligase (M2200; NEB) in 160 µl 1× quick ligation buffer (M2200; NEB) for 15 min at 25 °C. DNA was denatured at 95 °C for 2 min and cleaned with 1× AMPure XP beads. To generate UMI-4C libraries, two nested PCRs were performed using GoTaq polymerase (Promega) with a final primer concentration of 0.4 mM. The first PCR used the upstream bait primer (Supplementary Table 5) and Illumina universal primer 2 and amplification was performed for 20 cycles. The DNA was cleaned with 1× AMPure XP and used for the second PCR with the downstream bait primer (Supplementary Table 5) and Illumina universal primer 2 for 16 cycles. After the second PCR, the DNA was cleaned and size selected with 0.7× AMPure XP beads. The size distribution of the libraries was controlled by Bioanalyzer and libraries were quantified with a KAPA Quantification Kit (07960166001; Roche). Libraries were sequenced on a HiSeq 2500 using 2 × 125 bp reads or a NovaSeq S4 using 2 × 150 bp reads.
UMI-4C-seq analysis
Umi4cPackage (version 0.0.0.9000) was used as described43. FASTQ files from sequenced libraries were initially pooled by genotype. Paired-end reads were demultiplexed using fastq-multx from ea-utils (version 1.3.1). Reads were aligned and the number of UMIs extracted using p4cCreate4CseqTrack. A window of 1 kb around the viewpoint was removed from the analysis. 4C contact profiles from knockouts and controls were normalized for UMI coverage using the plotCompProf function and an adaptative smoothing method that controls window size so that no fewer than five molecules are included in each window. Assessment of differential contacts between knockout and wild-type 4C profiles in genomic regions of interest within a 0.5-megabase window surrounding the viewpoint was carried out using p4cIntervalsMean through a chi-squared test of normalized molecule counts.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The investigators were not blinded to allocation during the experiments and outcome assessment.
The results are shown as mean or median values, with error bars representing the s.e.m. or s.d., as stated in the figure captions. The numbers of biological replicates for each experiment are stated in the figure captions. P values were calculated by 𝜒2 test, two-sided Fisher’s exact test, unpaired two-tailed Student’s t-test or Wald or Wilcoxon rank-sum tests, as reported in the figure captions. The Brown–Forsythe test was used to test the equality of variances. For UMI-4C comparisons, P values were calculated by 𝜒2 test using umi4c. Statistical analysis of other epigenomic data is described in the appropriate Methods sections.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw sequence reads from RNA-seq, small conditional RNA-seq and ChIP-seq, as well as ChIP-seq peaks, are available from ArrayExpress under accession codes E-MTAB-11463, E-MTAB-11471, E-MTAB-11472, E-MTAB-11473, E-MTAB-11474, E-MTAB-11475 and E-MTAB-11477. The following previously published data were re-analysed: mouse liver ChIP-seq for CTCF (GSE29184; ref. 77), RAD21 (GSE102997; ref. 78), FOXA2, CEBPB and HNF4A (GSE57559; ref. 38), PPARA (GSE108689; ref. 79), RXR (GSE35262; ref. 80) and GATA4 (GSE49132; ref. 81); mouse liver and kidney ATAC-seq (SRP167062; ref. 82); and RNA-seq in humans (SRX218942; ref. 83), chickens (SRX2704301 ref. 84), X. tropicalis (SRX2704321; ref. 84), zebrafish (E-MTAB-8959; ref. 85), mouse kidneys (SRX2370375; ENCODE86), the small intestine (SRX2370402; ENCODE86); and human pluripotent stem cells differentiated into β cells (GSE140500; ref. 39). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
All of the custom code used in this study is available upon reasonable request.
References
Hon, C.-C. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015).
Gil, N. & Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117 (2020).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009).
Moran, I. et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16, 435–448 (2012).
Akerman, I. et al. Human pancreatic β cell lncRNAs control cell-specific regulatory networks. Cell Metab. 25, 400–411 (2017).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Ramilowski, J. et al. Functional annotation of human long non-coding RNAs via molecular phenotyping. Genome Res. 30, 1060–1072 (2020).
Anderson, K. M. et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436 (2016).
Cho, S. W. et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412 (2018).
Engreitz, J. M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016).
Wang, K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011).
Rom, A. et al. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat. Commun. 10, 5092 (2019).
Allou, L. et al. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature 592, 93–98 (2021).
Tronche, F. & Yaniv, M. HNF1, a homeoprotein member of the hepatic transcription regulatory network. Bioessays 14, 579–587 (1992).
Yamagata, K. et al. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3). Nature 384, 455–458 (1996).
Consortium, S. T. D. et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. J. Am. Med. Assoc. 311, 2305–2314 (2014).
Flannick, J., Johansson, S. & Njolstad, P. R. Common and rare forms of diabetes mellitus: towards a continuum of diabetes subtypes. Nat. Rev. Endocrinol. 12, 394–406 (2016).
Pontoglio, M. et al. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J. Clin. Invest. 101, 2215–2222 (1998).
Servitja, J. M. et al. Hnf1α (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol. Cell. Biol. 29, 2945–2959 (2009).
Shih, D. Q. et al. Loss of HNF-1α function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes 50, 2472–2480 (2001).
Lee, Y. H., Sauer, B. & Gonzalez, F. J. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α knockout mouse. Mol. Cell. Biol. 18, 3059–3068 (1998).
Ferrer, J. A genetic switch in pancreatic β-cells: implications for differentiation and haploinsufficiency. Diabetes 51, 2355–2362 (2002).
Pontoglio, M. et al. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84, 575–585 (1996).
Ding, C. H. et al. The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 17, 63 (2018).
Wang, C. et al. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed. Pharmacother. 89, 926–932 (2017).
Yang, X. et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 63, 881–890 (2014).
Zhan, Y. et al. Long non-coding RNA HNF1A-AS1 promotes proliferation and suppresses apoptosis of bladder cancer cells through upregulating Bcl-2. Oncotarget 8, 76656–76665 (2017).
Zhu, W. et al. Knockdown of lncRNA HNF1A-AS1 inhibits oncogenic phenotypes in colorectal carcinoma. Mol. Med. Rep. 16, 4694–4700 (2017).
Hannan, N. R., Segeritz, C. P., Touboul, T. & Vallier, L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 8, 430–437 (2013).
Parviz, F., Li, J., Kaestner, K. H. & Duncan, S. A. Generation of a conditionally null allele of hnf4α. Genesis 32, 130–133 (2002).
Boj, S. F., Parrizas, M., Maestro, M. A. & Ferrer, J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl Acad. Sci. USA 98, 14481–14486 (2001).
Fernandez Garcia, M. et al. Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75, 921–932 (2019).
Huang, P. et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 (2011).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Iwafuchi-Doi, M. et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 (2016).
Alvarez-Dominguez, J. R. et al. Circadian entrainment triggers maturation of human in vitro islets. Cell Stem Cell 26, 108–122 (2020).
Balboa, D. et al. Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. eLife 7, e38519 (2018).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 (2018).
Schwartzman, O. et al. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods 13, 685–691 (2016).
Ritter, N. et al. The lncRNA locus Handsdown regulates cardiac gene programs and is essential for early mouse development. Dev. Cell 50, 644–657 (2019).
Athineos, D. & Sansom, O. J. Myc heterozygosity attenuates the phenotypes of APC deficiency in the small intestine. Oncogene 29, 2585–2590 (2010).
McFadden, D. G. et al. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189–201 (2005).
Fernandez-Perez, A. et al. Hand2 selectively reorganizes chromatin accessibility to induce pacemaker-like transcriptional reprogramming. Cell Rep. 27, 2354–2369 (2019).
Miguel-Escalada, I., Pasquali, L. & Ferrer, J. Transcriptional enhancers: functional insights and role in human disease. Curr. Opin. Genet. Dev. 33, 71–76 (2015).
Osterwalder, M. et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Thorens, B. et al. Ins1Cre knock-in mice for beta cell-specific gene recombination. Diabetologia 58, 558–565 (2015).
Zhang, H., Fujitani, Y., Wright, C. V. & Gannon, M. Efficient recombination in pancreatic islets by a tamoxifen-inducible Cre-recombinase. Genesis 42, 210–217 (2005).
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA 93, 5860–5865 (1996).
Parrizas, M. et al. Hepatic nuclear factor 1-α directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol. Cell. Biol. 21, 3234–3243 (2001).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Miguel-Escalada, I. et al. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat. Genet. 51, 1137–1148 (2019).
Cebola, I. Deletion of regulatory elements with all-in-one CRISPR–Cas9 vectors. Methods Mol. Biol. 2351, 321–334 (2021).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Benazra, M. et al. A human beta cell line with drug inducible excision of immortalizing transgenes. Mol. Metab. 4, 916–925 (2015).
Miyazaki, J. et al. Establishment of a pancreatic β cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127, 126–132 (1990).
Pagliuca, F. W. et al. Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014).
Raj, A. & Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386 (2010).
Maestro, M. A. et al. Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum. Mol. Genet. 12, 3307–3314 (2003).
Bhatt, D. M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012).
Scotto-Lavino, E., Du, G. & Frohman, M. A. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2742–2745 (2006).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Keane, T. M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011).
Van de Geijn, B., McVicker, G., Gilad, Y. & Pritchard, J. K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 12, 1061–1063 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Cebola, I. et al. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 17, 615–626 (2015).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Hafemeister, C. & Satija, R.Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Matthews, B. J. & Waxman, D. J.Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. eLife 7, e34077 (2018).
Quagliarini, F. et al. Cistromic reprogramming of the diurnal glucocorticoid hormone response by high-fat diet. Mol. Cell 76, 531–545 (2019).
Boergesen, M. et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32, 852–867 (2012).
Zheng, R. et al. Function of GATA factors in the adult mouse liver. PLoS ONE 8, e83723 (2013).
Liu, C. et al. An ATAC-seq atlas of chromatin accessibility in mouse tissues. Sci. Data 6, 65 (2019).
Schultz, M. D. et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015).
Marin, R. et al. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 27, 1974–1987 (2017).
Gillard, G. B. et al. Comparative regulomics supports pervasive selection on gene dosage following whole genome duplication. Genome Biol. 22, 103 (2021).
Mouse, E. C. et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13, 418 (2012).
Acknowledgements
This research was supported by the Medical Research Council (MR/L02036X/1), Wellcome Trust (WT101033), European Research Council (Advanced Grant 789055), Spanish Ministry of Science and Innovation (BFU2014-54284-R and RTI2018-095666-B-I00) and National Institute for Health Research Imperial Biomedical Research Centre. Work in the Centre for Genomic Regulation was supported by the Centres de Recerca de Catalunya programme, Generalitat de Catalunya, Centro de Excelencia Severo Ochoa (CEX2020-001049) and by the Spanish Ministry of Science and Innovation aid to the European Molecular Biology Laboratory partnership. We thank the University of Barcelona School of Medicine animal facility, Center for Genomic Regulation and Imperial College London genomics units, Imperial College High Performance Computing Service, as well as K. Kaestner (University of Pennsylvania), M. Gannon (Vanderbilt University) and D. Tuveson (Cold Spring Harbor Laboratory) for the Cre lines. We thank J. Valcarcel and T. Graf (Center for Genomic Regulation) for comments on the manuscript, F. X. Real (Spanish National Cancer Research Centre) for insights and support, V. Grau and C. Roth for technical support and members of J.F.’s laboratory for valuable discussions.
Author information
Authors and Affiliations
Contributions
A. Beucher and J.F. conceived of, coordinated and supervised the study. A. Beucher performed the cell-based and computational studies and supervised the mouse analysis. A. Beucher, M.A.M. and J.G.-H. performed the analysis of mouse mutants. M.G.D.V. and P.R. designed and created the cell models. R.G.-F. performed the fluorescence in situ hybridization with supervision from A. Beucher. A. Beucher and I.M.-E. performed the UMI-4C studies. A. Beucher, A. Bernal and D.B. performed the stem cell studies. A. Beucher designed the mouse models with input from J.F., S.O. and P.V. S.O. and P.V. created the recombinant mice. H.H. participated in the single-cell genomics. A. Beucher and J.F. wrote the manuscript with input from the remaining authors.
Corresponding authors
Ethics declarations
Competing interests
P.R. is a founder and consultant for EndoCells/UniverCell Biosolutions. All of the remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Igor Ulitsky and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HASTER and HNF1A regulatory landscapes.
a, Chromatin and RNA maps in mouse (top) and human islets (bottom). Human CAGE is from islets, and mouse CAGE is from pancreas. The two mouse Haster transcriptional start sites are highlighted in blue, although only one transcriptional origin is apparent in human islets. The E islet enhancer, and CTCF-bound C region, both of which are bound by islet transcription factors, are highlighted in beige. b, HNF1A and HASTER expression across GTEx human tissues (Data Source: GTEx Analysis Release V8) and human islets (n = 130). Boxes show median and interquartile ranges. c, HASTER and HNF1A median transcript levels across tissues are negatively correlated, with the exception of whole pancreas.
Extended Data Fig. 2 HASTER transcripts localize to the nucleus.
a, Relative subcellular expression of HASTER lncRNA in EndoC-βH3 cells, compared to control mRNAs (TBP and HNF1A) and the nuclear lncRNA MALAT1. Mean ± s.d., n = 3 biological replicates. b, Single molecule fluorescence in situ hybridization signals for HASTER. HASTER transcripts are almost exclusively observed in the nucleus (deconvoluted images). The inset shows a rare non-nuclear signal. (n = 5 independent experiments). c, Colocalization of single molecule fluorescent in situ hybridization signals for HASTER (exonic probes) and HNF1A (intronic probes for HNF1A) in human EndoC-βH3 cells. n = 496 cells. The degree to which HASTER and intronic HNF1A RNA molecules are located at the HNF1A locus can only be assessed when two HNF1A or two HASTER molecules are seen in the same nucleus. In all such instances HNF1A and HASTER were found to colocalize. Scale bar, 20 µm.
Extended Data Fig. 3 Conditional Haster allele and phenotypic analysis in liver.
a, Schematic of the targeted allele, digestion fragments and probes used for Southern blot analysis of different alleles. b, Southern blot with KpnI (left) and NdeI (right) digestion. Asterisk, Clone 5 was selected to establish the line. n = 1 Southern blot. K, KpnI; N, NdeI; ES, parental embryonic stem cell (C57BL/6). c, Intraperitoneal glucose tolerance test in HasterLKO and control 8-week-old mice. Mean ± s.e.m. d, Body weight at 8 weeks for HasterLKO and controls. Mean ± s.d. c,d, n = 9 HasterLKO, n = 6 Alb-Cre;Haster+/+ and n = 7 Hasterf/f, two-tailed Student’s t-test. e, Immunofluorescence showing HNF1A overexpression in Haster-/- liver. n = 1 per genotype. Scale bar, 100 µM. f, GSEA displaying the enrichment of functional annotations in HasterLKO upregulated (top panel) and downregulated (bottom panel) genes.
Extended Data Fig. 4 HASTER is sensitive to decreased or increased HNF1A expression.
a, Locked nucleic acid (LNA) GapmeR knockdown of HNF1A (#1, HNF1A exon 8; #2, HNF1A exon 1) in human EndoC-βH3 β cells led to decreased HASTER RNA, and minor changes in other HNF1A-dependent genes. n = 3 nucleofections, TBP-normalized mean ± s.d.; two-tailed Student’s t-test. b, CRISPR-SAM activation of Hnf1a in mouse MIN6 β cells. n = 3 lentiviral transductions, representative of 2 independent experiments. Tbp-normalized mean ± s.d.; two-tailed Student’s t-test.
Extended Data Fig. 5 Hnf1a upregulation perturbs HNF1A binding selectivity.
a, Tissue specificity of gene expression across HNF1A-expressing tissues for genes upregulated in HasterLKO liver. To quantify tissue specificity, for each gene and tissue we calculated a Z-score that represents the deviation of expression in that tissue relative to the average from all tissues. n = 3 kidney samples, n = 5 liver and small intestine samples and n = 6 pancreatic islet samples. Box plots show medians and interquartile ranges; whiskers, 1.5 times the interquartile ranges. Two-sided Wilcoxon rank-sum P-values. b, Liver H3K4me3 and H3K27ac in HasterLKO and control liver (log2 normalized ChIP-seq read count; n = 3 mice per genotype). Red, differential H3K4me3 or H3K27ac sites (FDR ≤ 0.05); blue, kernel density of differential H3K4me3 or H3K27ac sites with FDR > 0.05. c, H3K27ac at HNF1A-bound regions in HasterLKO and controls (average of n = 3 mice per genotype). d, RNA fold change in HasterLKO vs. control liver of HNF1A-bound promoters for the different categories of HNF1A binding in HasterLKO liver. n = 5 mice per genotype for RNA and n = 3 mice per genotype for HNF1A ChIP. Box plots show medians and interquartile ranges; whiskers, 1.5 times the interquartile ranges. e, Examples of HNF1A neo-binding sites that lead to ectopic promoter and gene activation in HasterLKO liver. f, Ectopic activation of an intragenic promoter in HasterLKO liver (n = 2 mice per genotype). y axes in e and f represent MACS2 P values for ChIP-seq and RPKM for RNA-seq. g, Activated genomic regions that are bound by HNF1A and become active promoters in HasterLKO, but are inactive in control liver, overlap less frequently with annotated promoter and FANTOM5 CAGE transcriptional start sites, compared with unchanged HNF1A-bound active promoters in control liver, suggesting that some may be aberrant promoters rather that repurposed from other cell types. Two-sided Fisher’s exact test odd ratio (OR) and P-values, n = 3 mice per genotype.
Extended Data Fig. 6 Characterization of Haster mutants.
a, Body weight of males at 8 weeks of age. HasterpKO (n = 8), Pdx1-Cre;Haster+/+ (n = 12) and Hasterf/f (n = 8). Mean ± s.d. b, Intraperitoneal glucose tolerance test in 8-week-old and 30-week-old female HasterpKO (n = 9), Pdx1-Cre;Haster+/+ (n = 10) and Hasterf/f (n = 10). Mean ± s.e.m., *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 (two-tailed Student’s t-test). c, Body weight of 8- to 10-week-old male Haster-/- (n = 9), Haster+/- (n = 12) and Haster-/- (n = 13). Mean ± s.d. d, Intraperitoneal glucose tolerance test in 8- to 10-week-old females Haster-/- (n = 10), Haster+/- (n = 10) and Haster+/+ (n = 12). Mean ± s.e.m., *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 (two-tailed Student’s t-test). e, Relative quantification of HNF1A-negative β cells at the indicated age and genotype. Results show that HNF1A silencing correlates with time of Haster knockout, with higher silencing frequency after early deletion (Haster germline KO and HasterpKO models). HNF1A silencing increased with time in β cells from germline KO and HasterpKO models, but not when excision occurred in early β cells (βKO). No HNF1A silenced β cells were observed after Pdx1-CreERTM-based tamoxifen-inducible excision in adult β cells (HasteriβKO model). Mean ± s.d from 2-4 sections per mouse, 2-4 mice per condition. f-h, Immunofluorescence for HNF1A and insulin in E15.5 (f) Haster+/+, (g) HasterpKO, and (h) Haster-/- pancreas. Solid arrowheads, insulin cells overexpressing HNF1A; hollow arrowheads, insulin cells lacking HNF1A. i-k, Immunofluorescence for HNF1A and insulin in adult (i) wild-type, (j) HasterβKO and (k) HasteriβKO pancreas. Arrows point to HNF1A-negative β cells. Most β cells from HasterβKO and HasteriβKO islets overexpress HNF1A. e-k, n = 2 wild type embryos and n = 3 adult wild type mice; n = 2 Haster-/- embryos and adult mice; n = 3 HasterpKO embryos and adult mice; n = 2 HasterβKO 6- and 32-week-old mice; n = 2 HasteriβKO mice. n.a.: not analyzed because deletions were performed at a later time point. Scale bar, 50 µM.
Extended Data Fig. 7 Single-cell RNA-seq of HasterpKO islets.
a, Clusters of islet cells from female HasterpKO (4961 cells from triplicates for Seurat; 4456 for scVI) and controls (4646 cells from triplicates for Seurat; 4460 for scVI) determined by Seurat (t-SNE projection) or scVI (UMAP projection). b, scVI UMAP and Seurat t-SNE projections showing hormone expression. c, Cell type assignment based on marker gene expression. d, Haster and Hnf1a mRNA (log normalized UMI count) in different cellular populations of control and HasterpKO islets (Seurat). e, HNF1A-regulated gene expression (average Z-score) showing lower expression of HNF1A-regulated genes in the HASTERpKO-enriched β cell cluster (HNF1Alow) and high expression of HNF1A-regulated genes in other β cells in HASTERpKO cells. f, Relative proportions of β cells present in the HASTERpKO-enriched HNF1Alow β cell cluster. n = 3 mice per genotype. Mean ± s.d., two-tailed Student’s t-test. We note these proportions are lower than observed in situ, plausibly because Hnf1a-/- islets have a marked propensity to dissociate upon collagenase digestion, which is expected to cause negative selection of HNF1A-deficient cells after digestion and FACS sorting of single cells. g, HNF1A-regulated gene expression (average Z-score) for different cell types in individual samples (Seurat). h, Histograms showing the distribution of HNF1A-regulated gene expression (average Z-score) for β, α and δ cells (Seurat). Bins = 40. The variance of HNF1A-regulated gene expression increased in HasterpKO β, α and δ cells, showing that HNF1A-regulated genes are either upregulated or downregulated in islet cells. Levene test P-values.
Extended Data Fig. 8 Differential gene expression in HasterpKO β cells.
a, Genes differentially expressed in the major β-cell cluster of HasterpKO islets. Many of the most upregulated genes in HasterpKO islets are downregulated in Hnf1a-/- islets (blue horizontal lines). b, Examples of two genes that are known to be downregulated in Hnf1a-/- islets, Cpb2 and Gc, and show increased expression in HasterpKO β cells. c, GSEA showing upregulation in HasterpKO β cells of genes downregulated in Hnf1a KO islets. d, Genes that are downregulated (combined P ≤ 0.05) in HasterpKO HNF1Alow cells are often downregulated in Hnf1a-/- islets (blue horizontal lines). e, Expression of selected genes that are known to be downregulated in Hnf1a KO islets and are downregulated in HasterpKO HNF1Alow cells. Dots are medians of samples (log normalized UMI count) and bars are means of 3 replicates. Two-sided Wilcoxon Rank Sum test, P-values for the different biological replicates combined with Fisher’s method.
Extended Data Fig. 9 HASTER perturbations in β cells.
a,b, HASTER and HNF1A RNA in EndoC-βH3 cells with clonal homozygous deletion of (a) HASTER P1 promoter (HASTERΔP1/ΔP1) or (b) HASTER P1 and P2 promoters (HASTERΔP/ΔP). Deletion #1 and #2 were generated with independent pairs of sgRNAs, HASTER+/+ clones were transfected with sgRNAs targeting the AAVS1 locus. n = 4 clones per deletion. c, Two sets of LNA oligonucleotides (GapmeRs) were used to elicit HASTER degradation in EndoC-βH3 cells, without significant changes in HNF1A or HNF4A mRNA. n = 3 nucleofections. a-c, Expression normalized by TBP. Mean ± s.d., two-tailed Student’s t-test. d, Haster activation by CRISPR-SAM in MIN6 mouse β cells had no effect on Hnf1a expression. n = 3 lentiviral transductions. Expression normalized by Tbp. Mean ± s.d., two-tailed Student’s t-test relative to the control #1 sgRNA.
Extended Data Fig. 10 HNF1A binding to HASTER promoter reduces HNF1A promoter – enhancer interactions.
a, Top, UMI-4C profile trends using a viewpoint region upstream of Hnf1a (V1), near a CTCF-bound C site, in adult liver from n = 3 wild type (blue) or mutant (red) mice. Bottom, chromatin features and transcription factor binding in adult mouse liver. The Hnf1a upstream region contacts several enhancers, promoters and CTCF/cohesin sites in control and HasterLKO liver. The interaction between Hnf1a upstream region and E (asterisk) is increased in HasterLKO liver (see also Fig. 7). The region deleted in HasterLKO mice is highlighted in blue. b, UMI-4C profile trends of doxycycline-induced HNF1A overexpression in HASTER+ / + and HASTERΔP/ΔP EndoC-βH3 cells with HNF1A promoter as viewpoint. Strongest contacts occurred within < 20 kb 3’ of HNF1A promoter, while weaker contacts were predominantly observed in a ~400 kb region 5’ of HNF1A promoter. Top tracks, genes and regulatory elements in human pancreatic islets (Miguel-Escalada, et al., 2019). Pool of libraries for n = 4 independent experiments. c, Individual UMI-4C profile trends from four individual experiments of doxycycline-induced HNF1A overexpression in HASTER+ / + and HASTERΔP/ΔP EndoC-βH3 cells. In a-c shades represent estimated binomial standard deviation centered on the profile trend. d, HNF1A promoter – E interaction frequencies from individual replicates (n = 4 independent experiments). Interaction frequencies were measured at a 5 kb region centered on E highlighted with a green shade. Box plots show medians and interquartile ranges; whiskers, 1.5 times the interquartile ranges. e, Human islet chromatin marks showing the position of enhancers in the vicinity of HNF1A. f, HASTER+ / + or HASTERΔP1/ΔP1 clone #1 cells carrying targeted deletions in C (ΔC), E (ΔE) or sgGFP as control (WT). HASTER+ / + control and E deletion are identical to Fig. 7e. ΔC and ΔE were polyclonal deletions. Results are expressed as fold-differences relative to the parental HASTER+ / + or HASTERΔP1/ΔP1 cells. This showed that ΔC has no effect on HASTER or HNF1A, ΔE had significant effects on HASTER but did not significantly affect HNF1A in wild type cells, yet showed a significant HNF1A reduction in HASTERΔP1/ΔP1 cells. This is shown in cartoon form in the right panel, whereby E predominantly enhances HASTER transcription, but enhances HNF1A in the absence of HASTER. Pool of n = 3 independent experiments with 3 pairs of sgRNAs for each deletion. TBP-normalized mean expression ± s.d.; two-tailed Student’s t-test.
Supplementary information
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Beucher, A., Miguel-Escalada, I., Balboa, D. et al. The HASTER lncRNA promoter is a cis-acting transcriptional stabilizer of HNF1A. Nat Cell Biol 24, 1528–1540 (2022). https://doi.org/10.1038/s41556-022-00996-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00996-8
This article is cited by
-
Transcription regulation by long non-coding RNAs: mechanisms and disease relevance
Nature Reviews Molecular Cell Biology (2024)
-
The pancancer overexpressed NFYC Antisense 1 controls cell cycle mitotic progression through in cis and in trans modes of action
Cell Death & Disease (2024)
-
Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells
Nature Communications (2024)
-
Research progress and potential application of microRNA and other non-coding RNAs in forensic medicine
International Journal of Legal Medicine (2024)
-
Pancreatic microexons regulate islet function and glucose homeostasis
Nature Metabolism (2023)