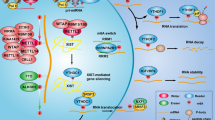
Abstract
METTL3 encodes the predominant catalytic enzyme to promote m6A methylation in nucleus. Recently, accumulating evidence has shown the expression of METTL3 in cytoplasm, but its function is not fully understood. Here we demonstrated an m6A-independent mechanism for METTL3 to promote tumour progression. In gastric cancer, METTL3 could not only facilitate cancer progression via m6A modification, but also bind to numerous non-m6A-modified mRNAs, suggesting an unexpected role of METTL3. Mechanistically, cytoplasm-anchored METTL3 interacted with PABPC1 to stabilize its association with cap-binding complex eIF4F, which preferentially promoted the translation of epigenetic factors without m6A modification. Clinical investigation showed that cytoplasmic distributed METTL3 was highly correlated with gastric cancer progression, and this finding could be expanded to prostate cancer. Therefore, the cytoplasmic METTL3 enhances the translation of epigenetic mRNAs, thus serving as an oncogenic driver in cancer progression, and METTL3 subcellular distribution can assist diagnosis and predict prognosis for patients with cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq, MeRIP–seq, RIP-seq and eCLIP–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO)45 under accession codes GSE163310 and GSE191170. The human cancer data were derived from the TCGA Research Network: http://cancergenome.nih.gov/. The dataset derived from this resource that supports the findings of this study is available in https://portal.gdc.cancer.gov/projects. Source data are provided with this paper.
References
He, P. C. & He, C. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40, e105977 (2021).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Su, R. et al. METTL16 exerts an mA-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24, 205–216 (2022).
Ping, X. L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Xu, W. et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 591, 317–321 (2021).
Schöller, E. et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24, 499–512 (2018).
Xie, J., Kozlov, G. & Gehring, K. The “tale” of poly(A) binding protein: the MLLE domain and PAM2-containing proteins. Biochimica et biophysica acta. 1839, 1062–1068 (2014).
Kranz, A. & Anastassiadis, K. The role of SETD1A and SETD1B in development and disease. Biochim Biophys Acta Gene Regul Mech 1863, 194578 (2020).
Fatscher, T., Boehm, V., Weiche, B. & Gehring, N. H. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 20, 1579–1592 (2014).
Umezawa, R. et al. Aberrant histone modifications at the thyrotropin-releasing hormone gene in resistance to thyroid hormone: analysis of F455S mutant thyroid hormone receptor. Endocrinology 150, 3425–3432 (2009).
Huang, J. et al. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol 80, 5740–5746 (2006).
Roguev, A. et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148 (2001).
Xue, H. et al. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 573, 445–449 (2019).
Berger, C. M., Gaume, X. & Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 113, 78–85 (2015).
Schick, M. et al. Genetic alterations of the SUMO isopeptidase SENP6 drive lymphomagenesis and genetic instability in diffuse large B-cell lymphoma. Nat. Commun. 13, 281 (2022).
Heerma van Voss, M. R. et al. Nuclear DDX3 expression predicts poor outcome in colorectal and breast cancer. OncoTargets Ther. 10, 3501–3513 (2017).
Vadlamudi, R. K. et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 65, 7724–7732 (2005).
Derry, J. J., Prins, G. S., Ray, V. & Tyner, A. L. Altered localization and activity of the intracellular tyrosine kinase BRK/Sik in prostate tumor cells. Oncogene 22, 4212–4220 (2003).
Bellovin, D. I., Bates, R. C., Muzikansky, A., Rimm, D. L. & Mercurio, A. M. Altered localization of p120 catenin during epithelial to mesenchymal transition of colon carcinoma is prognostic for aggressive disease. Cancer Res. 65, 10938–10945 (2005).
Tanaka, T. et al. Altered localization of retinoid X receptor alpha coincides with loss of retinoid responsiveness in human breast cancer MDA-MB-231 cells. Mol. Cell. Biol. 24, 3972–3982 (2004).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Huang, H. et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019).
Li, Y. et al. N6-methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 52, 870–877 (2020).
Niu, Y. et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics 11, 8–17 (2013).
Wei, G. et al. Acute depletion of METTL3 implicates N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome. Genome Res. 31, 1395–1408 (2021).
Wei, J. & He, C. Chromatin and transcriptional regulation by reversible RNA methylation. Curr. Opin. Cell Biol. 70, 109–115 (2021).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Cui, X., Meng, J., Zhang, S., Chen, Y. & Huang, Y. A novel algorithm for calling mRNA m6A peaks by modeling biological variances in MeRIP-seq data. Bioinformatics 32, i378–i385 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Quinlan, A. R. BEDTools: The Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47, 11–34 (2014). 11 12.
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lovci, M. T. et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 20, 1434–1442 (2013).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Acknowledgements
This work was funded by National Key Research and Development Program of China (2019YFA0801800 to J.Y., 2021YFA1102400 to F.W. and 2019YFA0111700 to X.W.); the National Natural Science Foundation of China (81530007 and 31725013 to J.Y., 82022001 and 81970103 to F.W., 82100135 to Y.G. and 82073129 to D.Z.); the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-019 to J.Y. and 2021-I2M-1-040 to F.W.); the Fundamental Research Funds for the Core Facility (3332019001), the CAMS (2016GH310001 to J.Y., 2017-I2M-B&R-04 to J.Y. and 2018RC310013 to F.W.) and the Medical Epigenetics Research Center, CAMS (2017PT31035); Outstanding Youths Development scheme of Nanfang Hospital, Southern Medical University (2020J001 to S.R.) and Outstanding Youths Development program, Southern Medical University (2019YQPY006 to S.R.). The open-access charge was funded by the National Key Research and Development Program of China (2019YFA0801800 to J.Y.).
Author information
Authors and Affiliations
Contributions
J.Y., F.W., J.J. and D.Z. conceived and designed the project. J.Y. and F.W. supervised the experiments. X.W. and M.H. prepared all the samples for next-generation sequencing. Y.H., Y.G. and Q.W. analysed all the data, and G.H. and J.X. helped to interpret data. J.P., Y.C. and D.L. performed the clinical and mouse experiments. X.W., J.P., M.H., P.S., L.X., H.W., W.S. and J.L. constructed all of the plasmids and stable cell lines, and performed the cell proliferation, migration and invasion assays, protein purification, western blotting, Co-IP, m6A quantification assay, MeRIP–qPCR, RIP–qPCR, IF and PLA. X.W. and Y.S. constructed cell fraction and polysome profiling. X.W. and Y.G conducted the in vitro transcription, in vitro translation and in vitro binding assays. X.W., Y.M. and S.Y. contributed with reagents and discussions. J.Y., F.W., S.R., X.W., Y.H. and Y.G. wrote and edited the manuscript, and all authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 METTL3 repression delays gastric tumorigenesis.
a, Correlation between METTL3 expression and perineural invasion (left panel) and between METTL3 expression and venous invasion (right panel) in GC patients (n = 113). The box plot indicates the median, the 25th and 75th percentiles (bounds of box), interquartile range, and the whiskers extending to the minima and maxima without considering outliers. b, Relative METTL3 expression in GC and normal tissues from the TCGA GC cohorts. c, Kaplan-Meier survival analysis of the overall survival of GC patients with high and low METTL3 expression from the TCGA GC cohorts. d, Heatmap of differential expressed genes between METTL3 high and low expressing groups. e, Gene ontology (GO) enrichment of 1,223 differential expressed genes in d. The bar chart shows the GO terms for pathways, ranked by -log10(p value). f, Somatic mutation patterns of GC patients from the TCGA dataset. The patients are divided into two groups based on METTL3 expression. g, Western blot analysis of METTL3 expression in MGC-803 cells (n = 3). h, Representative images of MGC-803-engrafted tumors treated with NTC or shMETTL3 at day 36 (n = 4). i, Representative images of H&E staining and immunostaining of METTL3, Ki67 and caspase-3 in the CDX model (n = 3). Scale bar, 100 μm. j, Representative images of PDX models treated with siMETTL3 or NTC (si-control) for 24 days (n = 4). Scale bar, 1 cm. k, Representative images of H&E staining and immunostaining of METTL3, Ki67 and caspase-3 in the PDX model (n = 3). Scale bar, 100 μm. The box plot (a) indicates the median (central mark), 25th and 75th percentiles (bounds of box), interquartile range, and the whiskers extending to the minima and maxima without considering outliers. The p values in (a, b) were determined by a Student’s t test (unpaired two-tailed). The p values in (c) were determined by log-rank with Mantel-Cox test. All data are shown as mean ± s.d.
Extended Data Fig. 2 METTL3 acts as an oncogene independently of its m6A methyltransferase activity.
a, Western blot analysis of METTL3 expression in METTL3_wt- or METTL3_cyto-transduced MGC-803 cells (left panel), and in METTL3_wt (middle panel) or METTL3_cyto (right panel) eCLIP precipitates (n = 3). b, A pie chart depicting the regional distribution of m6A sites identified by MeRIP-seq. c, Distribution of m6A locations across mRNA segments as identified by MeRIP-seq. d, Distribution of wild-type METTL3 binding sites across mRNA segments as identified by eCLIP-seq. e, The m6A peaks and METTL3 bound sites in KIAA1191, AASDHPPT and SUN1 transcripts belonging to the ‘Both’ group (upper panel). The m6A peaks in MYC, PKM and CDK1 transcripts belonging to the ‘m6A-only’ group (lower panel). The blue peaks represented the m6A peaks on the transcript, and the red peaks represented the accumulation of input. f, IF staining analysis of subcellular localization of METTL3 (green) in METTL3_wt and METTL3_mut-overexpressing MGC-803 and HGC-27 cells. Scale bar, 100 μm. g, The cellular proliferation rate of METTL3_wt- or METTL3_mut-transduced HGC-27 cells (n = 3). h, Transwell migration (left panel) and invasion (right panel) assays in METTL3_wt- or METTL3_mut-transduced HGC-27 cells (n = 3). i, Representative images of MGC-803-engrafted tumors; MGC-803 cells were treated with NTC or METTL3 constructs at day 36 (n = 4). The p values in (g, h) were determined by the Student’s t test (unpaired two-tailed). All data are shown as mean ± s.d.
Extended Data Fig. 3 Cytoplasmic METTL3 enables its oncogenic activity in GC cells.
a, Western blot analysis of fibrillarin and Hsp90 expression in cytoplasmic and nuclear fractions from MGC-803 cells treated with lysis buffer containing different NP-40 concentrations. From this result, 0.16% NP-40 (shown in the dotted box) was chosen for cytoplasmic and nuclear fractionation (n = 3). b, IF staining analysis of subcellular localization of METTL3 (green) in METTL3_cyto or METTL3_mut_cyto overexpressed MGC-803 and HGC-27 cells (n = 3). Scale bar, 100 μm. c, Western blot analysis of METTL3 expression in cytoplasmic and nuclear fractions from MGC-803 and HGC-27 cells treated by METTL3 sgRNA (M3 KO) and M3_wt or M3_cyto respectively (n = 3). d, Cell proliferation of MGC-803 and HGC-27 cells treated by different combination of METTL3 constructs as indicated (n = 3). e, Transwell migration (left panel) and invasion (right panel) assays of MGC-803 and HGC-27 cells treated by different combination of METTL3 constructs as indicated (n = 3). f, Cytoplasmic and nuclear METTL3 interactomes revealed by Co-IP analysis coupled with MS analysis. Proteins are sorted by ‘protein abundance’. Cytoplasmic PABPC1, and the nuclear partners WTAP and METTL14 are indicated by the red arrow. The p values in (d, e) were determined by the Student’s t test (unpaired two-tailed). All data are shown as mean ± s.d.
Extended Data Fig. 4 METTL3 promotes the translation of non-m6A-modified transcripts.
a, METTL3 distribution on different gene types revealed by wild-type (M3_wt) and the cytoplasmic (M3_cyto) METTL3 eCLIP-seq. b, The overlaps between m6A-modified mRNA detected by MeRIP-seq and METTL3-bound mRNA identified by the cytoplasmic METTL3 eCLIP-seq. c, Volcano plots of differentially expressed genes (DEGs) in METTL3_wt, METTL3_mut, shMETTL3 transcripts and their corresponding controls (NTC). Grey shading indicates genes with unchanged expression (log2 fold change < =1 and > = −1, or p > 0.05) compared with METTL3_wt, METTL3_mut, shMETTL3 transcripts and their corresponding controls (NTC). Red shading indicates up-regulated genes (log2 fold change>1 and p < 0.05) in METTL3_wt, METTL3_mut, shMETTL3 transcripts and their corresponding controls (NTC). Blue shading indicates down-regulated genes (log2 fold change < -1 and p < 0.05) in METTL3_wt, METTL3_mut, shMETTL3 transcripts and their corresponding controls (NTC). d, A Venn diagram illustrating target genes that are translationally activated by METTL3.
Extended Data Fig. 5 METTL3 interacts with PABPC1 to promote RNA looping.
a, Co-IP analysis using a METTL3-specific antibody under three concentrations of NP-40 (n = 3). b, Co-IP analysis using a METTL3-specific antibody in cells in which PABPC1 expression was knocked-down (n = 3).
Extended Data Fig. 6 METTL3 interacts with PABPC1 to promote the translation of oncogenic mRNAs.
a, The cellular proliferation rate of NTC or PABPC1-overexpressing MGC-803 cells (n = 3). b, Transwell cell migration (left panel) and invasion (right panel) assay of NTC or PABPC1-overexpressing MGC-803 cells (n = 3). c, The cellular proliferation rate of NTC or PABPC1-knockdown MGC-803 cells (n = 3). d, Transwell cell migration (left panel) and invasion (right panel) assay of PABPC1-knockdown and NTC MGC-803 cells (n = 3). e, Representative images of MGC-803-engrafted tumors in PABPC1-knockdown mice and PABPC1-competent control mice (day 36) (n = 4). f, Quantification of tumor growth (left panel) and tumor weight (right panel) in the CDX model following NTC or shPABPC1 treatment (n = 4). g, Left panel: rescue assay by transfection with NTC, PABPC1 (P1), shMETTL3 (shM3) or shMETTL3 (shM3) and PABPC1 (P1) (rescue) and western blot (n = 3). Right panel: Cellular proliferation rate of NTC, P1, shM3 or shM3 and P1 (rescue) MGC-803 cells. h, The number of migratory cells (left panel) and invading cells (right panel) of NTC, P1, shM3 or shM3 plus P1 (rescue) MGC-803 cells (n = 3). The p values in (a, b, c, d, f, g, h) were determined by the Student’s t test (unpaired two-tailed). All data are shown as mean ± s.d.
Extended Data Fig. 7 Cytoplasmic accumulation of METTL3 correlates with a poor outcome in GC and PC patients.
a, Relative METTL3 expression in GC and adjacent normal tissues. b, Left panel: correlations between total METTL3 expression and clinical stage in GC cancer patients. Right panel: Kaplan-Meier survival analysis of the overall survival probability of GC patients with low and high total METTL3 expression. c, Relative METTL3 expression in PC tissues and adjacent normal tissues. d, Left panel: the correlation between total METTL3 expression and clinical cancer stage in PC patients. Right panel: the correlation between METTL3 expression and Gleason grade in PC patients. e, Relative METTL3 expression in EC and adjacent normal tissues. f, Left panel: correlations between total METTL3 expression and clinical tumor stages in EC patients. Right panel: Kaplan-Meier survival analysis of the overall survival rates of EC patients with high and low METTL3 expression. g, Upper panels: correlations between the cytoplasmic to nuclear ratio of METTL3 and clinical tumor stages in EC patients. Lower panels: Kaplan-Meier survival analysis of overall survival rates of EC patients with high and low cytoplasmic to nuclear ratio of METTL3. h, Western blot analysis of METTL3 expression in cytoplasmic and nuclear fractions from DU145 (left panel) and KSYE510 (right panel) cells treated by M3_wt or M3_cyto respectively. i, Cell proliferation of DU145 (left panel) and KSYE510 (right panel) cells treated by different combination of METTL3 constructs as indicated (n = 3). j, Transwell migration and invasion assays of DU145 (upper panel) and KSYE510 (lower panel) cells treated by different combination of METTL3 constructs as indicated (n = 3). The p values in (a-g, i, j) for two-sample comparisons were determined by the Student’s t test (unpaired two-tailed). The p values in (b, f, g) for Kaplan-Meier survival analysis were determined by log-rank with Mantel-Cox test. For each violin in (b, d, f, g), the box plot indicates the median (white dot), 25th and 75th percentiles (bounds of box), interquartile range, and the whiskers extending to the minima and maxima without considering outliers. All data are shown as mean ± s.d.
Supplementary information
Supplementary Table 1
Supplementary Data Tables 1–9.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels.
Rights and permissions
About this article
Cite this article
Wei, X., Huo, Y., Pi, J. et al. METTL3 preferentially enhances non-m6A translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol 24, 1278–1290 (2022). https://doi.org/10.1038/s41556-022-00968-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00968-y
This article is cited by
-
METTL3 drives NSCLC metastasis by enhancing CYP19A1 translation and oestrogen synthesis
Cell & Bioscience (2024)
-
METTL3 promotes cellular senescence of colorectal cancer via modulation of CDKN2B transcription and mRNA stability
Oncogene (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
Exercise training decreases lactylation and prevents myocardial ischemia–reperfusion injury by inhibiting YTHDF2
Basic Research in Cardiology (2024)
-
METTL3-regulated m6A modification impairs the decidualization of endometrial stromal cells by regulating YTHDF2-mediated degradation of FOXO1 mRNA in endometriosis-related infertility
Reproductive Biology and Endocrinology (2023)