Abstract
Proliferation is a fundamental trait of cancer cells, but its properties and spatial organization in tumours are poorly characterized. Here we use highly multiplexed tissue imaging to perform single-cell quantification of cell cycle regulators and then develop robust, multivariate, proliferation metrics. Across diverse cancers, proliferative architecture is organized at two spatial scales: large domains, and smaller niches enriched for specific immune lineages. Some tumour cells express cell cycle regulators in the (canonical) patterns expected of freely growing cells, a phenomenon we refer to as ‘cell cycle coherence’. By contrast, the cell cycles of other tumour cell populations are skewed towards specific phases or exhibit non-canonical (incoherent) marker combinations. Coherence varies across space, with changes in oncogene activity and therapeutic intervention, and is associated with aggressive tumour behaviour. Thus, multivariate measures from high-plex tissue images capture clinically significant features of cancer proliferation, a fundamental step in enabling more precise use of anti-cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Images, generated datasets and corresponding analysis are available in the Synapse.org repository syn2230077174. Multiplexed images of human HER2 breast cancer used in Fig. 3a can be viewed in Minerva Story72,73 an interpretive guide for interacting with multiplexed tissue imaging data https://tinyurl.com/minerva-proliferation. Imaging data and other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
MATLAB codes used to process and analyse the p-CyCIF and t-CyCIF (shortened as ‘CyCIF’) and the codes used for ccD-CMD analysis are available at https://github.com/santagatalab.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Duval, K. et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology 32, 266–277 (2017).
Norton, L. A Gompertzian model of human breast cancer growth. Cancer Res. 48, 7067–7071 (1988).
Norton, L., Simon, R., Brereton, H. D. & Bogden, A. E. Predicting the course of Gompertzian growth. Nature 264, 542–545 (1976).
Collins, V. P., Loeffler, R. K. & Tivey, H. Observations on growth rates of human tumors. Am. J. Roentgenol. Radium Ther. Nucl. Med 76, 988–1000 (1956).
Black, J. R. M. & McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer https://doi.org/10.1038/s41568-021-00336-2 (2021).
Nia, H. T., Munn, L. L. & Jain, R. K. Physical traits of cancer. Science 370, eaaz0868 (2020).
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Nirmal, A. J. et al. The spatial landscape of progression and immunoediting in primary melanoma at single cell resolution. bioRxiv https://doi.org/10.1101/2021.05.23.445310 (2021).
Aguirre-Ghiso, J. A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 7, 834–846 (2007).
Inwald, E. C. et al. Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res Treat. 139, 539–552 (2013).
Lehr, H.-A. et al. Mitotic figure counts are significantly overestimated in resection specimens of invasive breast carcinomas. Mod. Pathol. 26, 336–342 (2013).
Cuylen-Haering, S. et al. Chromosome clustering by Ki-67 excludes cytoplasm during nuclear assembly. Nature 587, 285–290 (2020).
Sobecki, M. et al. The cell proliferation antigen Ki-67 organises heterochromatin. eLife 5, e13722 (2016).
Bruno, S. & Darzynkiewicz, Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 25, 31–40 (1992).
Miller, I. et al. Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 24, 1105–1112.e5 (2018).
Gerdes, J., Schwab, U., Lemke, H. & Stein, H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J. Cancer 31, 13–20 (1983).
Denkert, C. et al. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast 24, S67–S72 (2015).
Nielsen, T. O. et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in Breast Cancer Working Group. J Natl Cancer Inst https://doi.org/10.1093/jnci/djaa201 (2020).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Bodenmiller, B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2, 225–238 (2016).
Gookin, S. et al. A map of protein dynamics during cell-cycle progression and cell-cycle exit. PLoS Biol. 15, e2003268 (2017).
Mahdessian, D. et al. Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature 590, 649–654 (2021).
Nishitani, H., Lygerou, Z., Nishimoto, T. & Nurse, P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625–628 (2000).
McGarry, T. J. & Kirschner, M. W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053 (1998).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Kafri, R. et al. Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature 494, 480–483 (2013).
Gut, G., Tadmor, M. D., Pe’er, D., Pelkmans, L. & Liberali, P. Trajectories of cell-cycle progression from fixed cell populations. Nat. Methods 12, 951–954 (2015).
Cannoodt, R. et al. SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. bioRxiv https://doi.org/10.1101/079509 (2016).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
Liang, S., Wang, F., Han, J. & Chen, K. Latent periodic process inference from single-cell RNA-seq data. Nat. Commun. 11, 1441 (2020).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Takasaki, Y., Deng, J. S. & Tan, E. M. A nuclear antigen associated with cell proliferation and blast transformation. J. Exp. Med. 154, 1899–1909 (1981).
Bravo, R., Frank, R., Blundell, P. A. & Macdonald-Bravo, H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 326, 515–517 (1987).
Chong, J. P., Mahbubani, H. M., Khoo, C. Y. & Blow, J. J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 375, 418–421 (1995).
Madine, M. A., Khoo, C. Y., Mills, A. D. & Laskey, R. A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature 375, 421–424 (1995).
Cayrol, C., Knibiehler, M. & Ducommun, B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene 16, 311–320 (1998).
Sherr, C. J. & Roberts, J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 (1999).
Viale, G. et al. Prognostic and predictive value of centrally reviewed Ki-67 labeling index in postmenopausal women with endocrine-responsive breast cancer: results from Breast International Group Trial 1-98 comparing adjuvant tamoxifen with letrozole. J. Clin. Oncol. 26, 5569–5575 (2008).
Allegra, C. J. et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes’ B and C colon cancer: a National Cancer Institute–National Surgical Adjuvant Breast and Bowel Project collaborative study. J. Clin. Oncol. 21, 241–250 (2003).
DeSantis, C. E. et al. Breast cancer statistics, 2019. CA Cancer J. Clin. 69, 438–451 (2019).
Rakha, E. A. et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 12, 207 (2010).
Allred, D. C. et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J. Natl Cancer Inst. 85, 200–206 (1993).
Tannock, I. F. The relation between cell proliferation and the vascular system in a transplanted mouse mammary tumour. Br. J. Cancer 22, 258–273 (1968).
Zaidi, M., Fu, F., Cojocari, D., McKee, T. D. & Wouters, B. G. Quantitative visualization of hypoxia and proliferation gradients within histological tissue sections. Front. Bioeng. Biotechnol 7, 397 (2019).
Muthuswamy, S. K. Self-organization in cancer: implications for histopathology, cancer cell biology, and metastasis. Cancer Cell 39, 443–446 (2021).
Arata, Y. & Takagi, H. Quantitative studies for cell-division cycle control. Front Physiol. 10, 1022 (2019).
Lin, J.-R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Álvarez-Fernández, M. & Malumbres, M. Mechanisms of sensitivity and resistance to CDK4/6 inhibition. Cancer Cell 37, 514–529 (2020).
Hafner, M. et al. Multiomics profiling establishes the polypharmacology of FDA-approved CDK4/6 inhibitors and the potential for differential clinical activity. Cell Chem. Biol. 26, 1067–1080.e8 (2019).
Moasser, M. M. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26, 6469–6487 (2007).
Goel, S. et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell 29, 255–269 (2016).
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 36, 2105–2122 (2018).
Slamon, D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987).
Filho, O. M. et al. HER2 heterogeneity as a predictor of response to neoadjuvant T-DM1 plus pertuzumab: results from a prospective clinical trial. J. Clin. Oncol. 37, 502–502 (2019).
Katayama, A. et al. Predictors of pathological complete response to neoadjuvant treatment and changes to post-neoadjuvant HER2 status in HER2-positive invasive breast cancer. Mod. Pathol. https://doi.org/10.1038/s41379-021-00738-5 (2021).
Chittajallu, D. R. et al. In vivo cell-cycle profiling in xenograft tumors by quantitative intravital microscopy. Nat. Methods 12, 577–585 (2015).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Moser, J., Miller, I., Carter, D. & Spencer, S. L. Control of the restriction point by Rb and p21. Proc. Natl Acad. Sci. USA 115, E8219–E8227 (2018).
Min, M., Rong, Y., Tian, C. & Spencer, S. L. Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science 368, 1261–1265 (2020).
Amin, A. D., Rajan, S. S., Groysman, M. J., Pongtornpipat, P. & Schatz, J. H. Oncogene overdose: too much of a bad thing for oncogene-addicted cancer cells. Biomark. Cancer 7, 25–32 (2015).
Briscoe, J. & Small, S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development 142, 3996–4009 (2015).
Rajewsky, N. et al. LifeTime and improving European healthcare through cell-based interceptive medicine. Nature 587, 377–386 (2020).
HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Caswell-Jin, J. L. et al. Clonal replacement and heterogeneity in breast tumors treated with neoadjuvant HER2-targeted therapy. Nat. Commun. 10, 657 (2019).
Lomakin, A. et al. Spatial genomics maps the structure, character and evolution of cancer clones. bioRxiv https://doi.org/10.1101/2021.04.16.439912 (2021).
Rueda, O. M. et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 567, 399–404 (2019).
Zahir, N., Sun, R., Gallahan, D., Gatenby, R. A. & Curtis, C. Characterizing the ecological and evolutionary dynamics of cancer. Nat. Genet. 52, 759–767 (2020).
Hoffer, J. et al. Minerva: a light-weight, narrative image browser for multiplexed tissue images. J. Open Source Softw. 5, 2579 (2020).
Rashid, R. et al. Narrative online guides for the interpretation of digital-pathology images and tissue-atlas data. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-021-00789-8 (2021).
Gaglia, G. et al. Temporal and spatial topography of cell proliferation in cancer. Nat. Cell Biol. https://doi.org/10.7303/syn22300771 (2022).
Mills, C. E. et al. Multiplexed and reproducible high content screening of live and fixed cells using the Dye Drop method. bioRxiv https://doi.org/10.1101/2021.08.27.457854 (2021).
Du, Z. et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat. Protoc. 14, 2900–2930 (2019).
Peng, T. et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8, 14836 (2017).
Muhlich, J., Chen, Y.-A., Russell, D. & Sorger, P. K. Stitching and registering highly multiplexed whole slide images of tissues and tumors using ASHLAR software. bioRxiv https://doi.org/10.1101/2021.04.20.440625 (2021).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Creed, J. H., Gerke, T. A. & Berglund, A. E. MatSurv: survival analysis and visualization in MATLAB. J. Open Source Softw. 5, 1830 (2020).
Csikász-Nagy, A., Battogtokh, D., Chen, K. C., Novák, B. & Tyson, J. J. Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys. J. 90, 4361–4379 (2006).
Acknowledgements
This work was supported by NIH grants R01-CA194005 (S.S.), R41-CA224503 (P.K.S.), U54-CA225088 (P.K.S. and S.S.), P50-CA168504 (J.J.Z. and E.P.W.), R35-CA210057 (J.J.Z.), T32-GM007748 (S.C.), K08-CA191058 (R.J.) and T32-HL007627 (G.G.); DF/HCC Breast SPORE: Specialized Program of Research Excellence (SPORE), the Ludwig Center at Harvard (P.K.S. and S.S.), the American-Italian Cancer Foundation postdoctoral fellowship (G.G.), the Terri Brodeur Breast Cancer Foundation and BCRF-AACR Career Development Awards for Translational Breast Cancer Research (S.K.), the Breast Cancer Research Foundation BCRF (J.J.Z.), DOD CDMRP W81XWH-18-1-0491 (J.J.Z.), the Gray Foundation, and the BWH President’s Scholar Award (S.S.). We thank Dana-Farber/Harvard Cancer Center for the use of the Specialized Histopathology Core, which provided histopathology services. Dana-Farber/Harvard Cancer Center is supported in part by a National Cancer Institute (NCI) Cancer Center Support Grant P30-CA06516. Tissue samples were also provided by the Cooperative Human Tissue Network (CHTN), which is funded by the NCI. Other investigators may have received specimens from the same subjects. We thank C. Yapp and Y.-A. Chen for assistance with microscopy and image analysis, and C. Rycroft and G. Lahav for helpful discussions.
Author information
Authors and Affiliations
Contributions
G.G., S.K., J.J.Z., P.K.S. and S.S. were responsible for study conceptualization. G.G., S.K., S.W., C.E.M., J.B., S.C., J.R.L., P.K.S. and S.S. developed and implemented the methodology. G.G., S.K., D.R., Y.D., A.V., C.E.M., .M.C. and S.C. conducted data acquisition. G.G., D.R. and Y.D. generated software. G.G., S.K., D.R., Y.D., S.C., D.A.D. and D.R. performed data validation. G.G., S.K., D.R., Y.D., A.V., C.E.M. and S.C. performed formal analysis. J.B., D.A.D., R.J., O.M. and E.P.W. provided resources. G.G., S.K., D.R., Y.D., S.C. and D.A.D. performed data curation. G.G., S.K. and S.S. wrote the original draft, and all authors reviewed and edited the manuscript. Data visualization was conducted by G.G., S.K., Y.D. and D.R. J.J.Z., P.K.S. and S.S. provided project supervision, and P.K.S. and S.S. provided project administration. E.P.W., J.J.Z., P.K.S. and S.S. provided resources and funding.
Corresponding authors
Ethics declarations
Competing interests
P.K.S. is a member of the Scientific Advisory Board of RareCyte and NanoString Technologies, is on the Board of Directors of Applied Biomath and is a consultant to Merck and Montai Health. P.K.S. is co-founder of Glencoe Software, which contributes to and supports the open-source OME/OMERO image informatics software used in this paper. S.S. is a consultant for RareCyte, and J.J.Z. is a founder and board director of Crimson Biotech and Geode Therapeutics. J.S.B. is a scientific consultant and has stock options for Geode Therapeutics. D.A.D. is on the Advisory Board for Oncology Analytics and has consulted for Novartis. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Tobias Meyer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
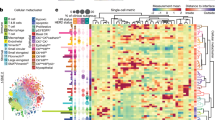
Extended Data Fig. 1 Multivariate Proliferation Index (MPI) in human cancer tissues.
a. Clustered heat map of log2 normalized cell lineage marker signal intensities on a per-cell basis derived from CyCIF images of 4 whole slides of lung squamous cell carcinoma (SCC) and ovarian carcinoma, and tissue microarrays (TMAs) from glioma, colorectal carcinoma (CRC) and mesothelioma. b-c. Clustered heat map of single-cell signal intensities of cell cycle markers for epithelial/tumour cells in Fig. 1a (breast carcinoma) and panel a. Ki-67+ cells were identified by normalization using Gaussian mixture modeling with 2 components. Multivariate Proliferation Index (MPI) indicated: +1 (proliferative, green), 0 (non-proliferative, blue), or -1 (arrested, red). d. K-means clustering heat map of five MPI markers for epithelial/tumour cells of three HER2 + breast cancer samples (k = 20 clusters), and heat map of single-cell normalized log2 intensities. In both, the corresponding MPI category is depicted for comparison (k-mean clustering fraction of MPI category is depicted). e. UMAP plots for 180 samples of breast carcinomas from the Cooperative Human Tissue Network (CHTN) Stage II TMAs (#14-17) with proliferation markers mapped to colour (MPI categories were not used as UMAP variables, n = 10,000 cells). f. MPI robustness comparison between five sets of serially cut tissue sections from Pantomics TMA BRC15010 and g. between two sets of serially cut tissue sections of the 3 breast HER2-positive cases from Fig. 1c,e. Each dot represents the fraction of MPI + 1 cells in the two indicated tissue sections (linear least-square fit with fixed origin at y = x = 0). h-j. Permutation testing of breast carcinomas from the CHTN Stage II TMAs (#14-17, n = 180 samples) with MPI calculations performed by adding increasing amounts of normally distributed noise (0.1x, 0.25x, 0.5x and 1x standard deviations of original marker distribution) to the five MPI cutoffs, repeated 1000 times using i. linear regression, i. correlation analysis (boxplots: median, 25-75th percentile and min-max extremes from MatLab boxplot.m function), and j. intra-cohort ordering of MPI estimates. k. Comparison of MPI 0, and MPI -1 fractions in epithelial/tumour cells across different classifiers of breast cancer in Fig. 1g (n = 142 samples, mean + SD, 2-sided KS p-values with 0.05 significance cutoff).
Extended Data Fig. 2 Multivariate Proliferation Index (MPI) short- and long-range correlations in human cancer tissues.
a-b. Composite CyCIF image from a. lung squamous cell carcinoma, and b. ovarian carcinoma (scale bar, 1 mm) and corresponding image of long-range whole slide spatial maps of MPI categories (smoothed over 40 neighbouring cells for visualization purposes only). Inset panel showing both smoothed and single-cell MPI calling in marked region of interest. Further inset panel of single-cell MPI calling and corresponding composite CyCIF image (scale bar, 100 µm; white = pan-cytokeratin, green = MCM2, red = p27, blue = DNA; example from 4 biological independent specimens). c. Spatial maps of MPI categories from inset 1 from Fig. 2a smoothed over 40 neighbouring cells and non-smoothed (single-cell calling). d. Red density curve shows sample distribution of spatial correlations within and across (“self corr” and “cross corr”) MPI categories for mesothelioma, glioma, colorectal carcinoma (CRC) and three breast TMAs (n = 52, 163, 89, 69, 85, 57 samples respectively, k = 5th neighbour approximation, kernel smoothing (KS) density approximation). Blue density curve shows bootstrap distribution comparison obtained by randomly shuffling of MPI labels (10 independent shuffles). e. Heat map of spatial correlations within and across randomly shuffled MPI labels (“self corr” and “cross corr”, k = 5th neighbour, n = 513 samples). f. 2-sided t-test p-values for red density curve distributions in panel d (log10 scale used for visualization purposes). g. Plots of spatial correlation from whole slides images of samples in Fig. 2d with two exponential fit for the three MPI categories. h. Ripley’s L-r function for MPI + 1 across breast, lung, and ovarian tumours (whole slide images, Fig. 2d). Each column represents an individual and independent tissue.
Extended Data Fig. 3 Limits of cell cycle marker single cell distributions and multi-channel gating interpretation in tissues.
a. Representative single-channel cell cycle marker images from CyCIF imaging with E-cadherin from tissue in Fig. 3a (scale bar 100 µm, example from 3 biological independent specimens). b. Single-channel distributions and two-dimensional scatter plots of a subset of cell cycle markers from HER2 positive breast cancer case #2 in Fig. 1c,e (n = 1,000 epithelial/tumour cells, log2 signal per cell prior to normalization). c. Single channel distributions for cell lineage, proliferation, and cell cycle markers from three HER2 + breast cancer samples (kernel smoothing (KS) density approximation) used in Fig. 1c,e. For proliferation and cell cycle markers only epithelial/tumour cells were used. d. Upset plot of three HER2 + breast cancer samples (n = 0.52 million cells) from Fig. 1c, e showing frequency of cell cycle marker positivity and their combinations sorted by frequency.
Extended Data Fig. 4 Example of 27-plex CyCIF experiment with tumour sample and tonsil control tissues.
a. Example of positive and negative staining for all markers in CyCIF experiment through rounds of cyclic imaging. Three independent samples are shown: Ctrl is a non-malignant tonsil tissue sample (‘control’), #1 and #2 are glioma samples. Plots are single-cell kernel smoothing (KS) density estimation for patient samples from respective images (median per pixels within the cell area, log2 FAU, not normalized, black = tonsil, yellow = glioma sample #1, magenta = glioma sample #2, n = 4,278, 2,629, and 2,609 cells, respectively). Each row of images and data is a successive round of CyCIF acquired from the same tissue area (Rx is the xth round of imaging). All images from antibody channels were linearly contrasted between 0 and 2000 fluorescence units for ease of comparison. Scale bar, 50 µm. Examples from tissue microarray containing 176 independent glioma specimens and 8 independent control specimens. b. Scatter plots of the single-cell correlation of the signal intensity of unconjugated antibodies to the indicated markers versus their fluorophore conjugated versions from 142 samples of tissue from 75 patients (Pantomics TMA BRC15010). Pearson correlation coefficients (C) are shown. All correlation had p-value < 10-175. c. Plot of the phospho-Rb signal from MPI0, MPI + 1/Ki-67-, and MPI + 1/Ki-67+ cells from Pantomics TMA BRC15010 acquired using both conjugated (‘direct IF’) and unconjugated (‘indirect IF’) phospho-Rb antibodies (n = 74 cores, Line is mean value).
Extended Data Fig. 5 Testing of ccD-CMD framework using cell cycle perturbation in vitro and cell cycle modeling in silico, and comparison of temporal inference methods.
a. Left, histograms of fluorescence single-cell log2 normalized signal by plate-based CyCIF (p-CyCIF) from untreated (blue) and 24 h palbociclib 1 µM treated (orange) MCF10A cells grown in culture (n = 10,000 cells per condition). Right, ccD-CMD plot. b. Heat map and time plot of single-cell signals ordered by ccD-CMD (normalized log2 FAU, n = 10,000 cells moving mean over 200 cells). c. Traces from generalized model of mammalian cell cycle80. Top panels, no noise added. Middle panels, Gaussian measurement noise added (additive and multiplicative). Gray areas are the time frame used for plots in panels d. d. ccD-CMD plot of mathematical model results in shaded areas of panel c. Bottom panel, simulation switch to G1 arrest parameters (CDK/CyclinD complex set to 0) after one cell cycle. Colour is the time variable in the mathematical model (n = 10,000 points). e-h. Comparison of time inference methods ccD-CMD, SCORPIUS, Palantir and Cyclum applied to e. synthetic data generated by the mathematical model in panel e. (n = 10,000 observations), f. HER2 + breast cancer tissue data from Fig. 3a-f., and g. MCF10A untreated cells from Fig. 3g. Left, two dimensional visualization output. Right, pseudotime ordering output (n = 5,000 cells, same cells used for all algorithms, normalized log2 FAU, moving mean over 200 cells). h. Comparison between two dimensional reduced space visualization from three time inference algorithms with data from HER2 + breast patient samples #1, 2, and 3 from Fig. 4b-d. i. Coefficient of variation of coherence metrics IOV and CFD in 5 tissues using increasing number of cells (n > 20,000 cells per tissue, CV calculated over 40 independent sub-samplings). j. Comparison of IOV and CFD when one or two markers are removed from ccD-CMD algorithm. Data from untreated MCF10A cells used in panel g. The green dot is the original representation. Orange, one marker removed. Blue, two markers removed. k. Comparison of IOV and CFD when MPI 0 and MPI -1 cells are added at increasing ratios. l. Examples of pseudotime ordering by ccD-CMD algorithm showing inferred CDT1 and Geminin across six tissues areas.
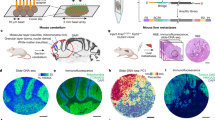
Extended Data Fig. 6 Orthogonal validation of changes in MPI and cell cycle coherence using cell cycle inhibition in vitro and in vivo.
a-b. Stacked bar graphs showing a. cell cycle fractions using the dye drop method75 and b. MPI frequencies by p-CyCIF for MCF-7 cells at baseline and in response indicated treatments for 24 h c. Cell cycle fractions in five breast cancer cell lines showing differential response to treatment with increasing doses of Palbociclib dependent on the Rb status of the cell line. d. ccD-CMD plot from p-CyCIF from fixed untreated cells (Control), and cells exposed to 1 µM Palbociclib for 24 h. Enlarged dot represents the average of 3 distinct biological replicates, n = 1000 cells each. e-g. t-CyCIF validation in MCF-7 xenografts in nude mice53. Doses are in mg/kg. e. Scatter plot of change in tumour size and MPI + 1 fraction. Vehicle, n = 14 tumours from 7 mice. Abemaciclib, n = 6 tumours from 3 mice per dose. Palbociclib, n = 8 tumours from 5 mice. Each dot represents an individual tumour (2-sided Pearson correlation p-value). f. MPI and Ki-67 fractions per TMA core (mean of n = 35, 15, 17, 18, and 24 total cores, 2-sided t-test p-value). g. ccD-CMD plots from individual MCF-7 xenografts tumours (4000 cells per mouse, n = 12, 6, 6, 6, 8 mice).
Extended Data Fig. 7 Cell cycle dynamics from ccD-CMD from human cancer tissues.
a. Scatter plot of CFD versus IOV (for each ROI from Fig. 4f). b. K-means clustering of cell cycle markers from selected ROIs used in Fig. 4g (n = 3600 cells per ROI, k = 15 clusters). Left, single-cell clustering with ROI annotation (log2 normalized FAU). Middle, cluster median. Right, ROI composition for each cluster. c. Scanned image of hematoxylin and eosin (H&E) stained section from three HER2 positive breast tissues with pathology annotations. d. Composite CyCIF images of tissues from panel c (green CD45, red Vimentin, white E-Cadherin, scale bar 2 mm). e. Inter-Octile Variation IOV, f. Circle Fit Distance (CFD), and g. MPI + 1 fraction for each ROI noted in panel d.
Extended Data Fig. 8 HER2 expression and cell cycle dynamics in human breast cancer tissues and HER2 driven mouse model of breast cancer.
a. ccD-CMD plots for two HER2 + breast cancer tissue microarrays (TMA # 1 and #2) for increasing levels for HER2 protein. Single cells binned by normalized HER2 levels (n = 5,000 cells per bin were used). Right, ccD-CMD scatter plots of highest HER2 bin for both TMA1 and 2 with single marker normalized intensities mapped to colour (n = 5,000 cells). b. ccD-CMD scatter plots of the single-cell data from MPI + 1 cells from time course of HER2 induction and repression in GEMM with single marker normalized intensities mapped to colour (n = 5,000 cells per plot, p27 was not used by the ccD-CMD algorithm). c. Mean + /- 25th percentile of circle fit distance of cell cycle markers in tumour cells (in situ and invasive) from Fig. 5h (n = 5,000 cells per plot).
Extended Data Fig. 9 Association between with clinical outcome and cell proliferation metrics.
a. Scatter plots of CFD versus IOV from mesothelioma and glioblastoma sample cohorts (n = 22 and 32 patients, respectively). Colors represent binning into coherence groups according IOV and CFD metrics. b-c. Kaplan Meier estimation and logrank p-value of progression-free survival (PFS) for the two patient cohorts in Fig. 7. Patients were binned in b. three groups “coherent”, IOVhigh “skewed” and IOVlow CFDhigh “non-canonical” groups from panel a). or c. two groups based on the median Ki-67+ fraction.
Supplementary information
Supplementary Information
Supplementary Note/Discussion
Supplementary Table 1
This file contains Supplementary Tables 1–8 as outlined in the main text.
Rights and permissions
About this article
Cite this article
Gaglia, G., Kabraji, S., Rammos, D. et al. Temporal and spatial topography of cell proliferation in cancer. Nat Cell Biol 24, 316–326 (2022). https://doi.org/10.1038/s41556-022-00860-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00860-9
This article is cited by
-
Multiplex protein imaging in tumour biology
Nature Reviews Cancer (2024)
-
Multi-omics pan-cancer analyses identify MCM4 as a promising prognostic and diagnostic biomarker
Scientific Reports (2024)
-
Qualification of a multiplexed tissue imaging assay and detection of novel patterns of HER2 heterogeneity in breast cancer
npj Breast Cancer (2024)
-
Spatial biology of cancer evolution
Nature Reviews Genetics (2023)
-
Targeting pan-essential pathways in cancer with cytotoxic chemotherapy: challenges and opportunities
Cancer Chemotherapy and Pharmacology (2023)