Abstract
Gastric cancer is among the most prevalent and deadliest of cancers globally. To derive mechanistic insight into the pathways governing this disease, we generated a Claudin18-IRES-CreERT2 allele to selectively drive conditional dysregulation of the Wnt, Receptor Tyrosine Kinase and Trp53 pathways within the gastric epithelium. This resulted in highly reproducible metastatic, chromosomal-instable-type gastric cancer. In parallel, we developed orthotopic cancer organoid transplantation models to evaluate tumour-resident Lgr5+ populations as functional cancer stem cells via in vivo ablation. We show that Cldn18 tumours accurately recapitulate advanced human gastric cancer in terms of disease morphology, aberrant gene expression, molecular markers and sites of distant metastases. Importantly, we establish that tumour-resident Lgr5+ stem-like cells are critical to the initiation and maintenance of tumour burden and are obligatory for the establishment of metastases. These models will be invaluable for deriving clinically relevant mechanistic insights into cancer progression and as preclinical models for evaluating therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE184613. Previously published TCGA data that were re-analysed here are available from the Broad Institute TCGA Genome Data Analysis Centre Firehose (https://gdac.broadinstitute.org/). The data that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
Code will be made available upon reasonable request
References
GLOBOCAN Cancer Fact Sheet (WHO International Agency for Cancer Research, 2019).
Rahman, R., Asombang, A. W. & Ibdah, J. A. Characteristics of gastric cancer in Asia. World J. Gastroenterol. 20, 4483–4490 (2014).
Tan, P. & Yeoh, K. G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology 149, 1153–1162.e3 (2015).
Lochhead, P. & El-Omar, E. M. Gastric cancer. Br. Med. Bull. 85, 87–100 (2008).
Goldenring, J. R. & Nam, K. T. Oxyntic atrophy, metaplasia, and gastric cancer. Prog. Mol. Biol. Transl. Sci. 96, 117–131 (2010).
Goldenring, J. R., Nam, K. T., Wang, T. C., Mills, J. C. & Wright, N. A. Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology 138, 2207–2210.e1 (2010).
Grabsch, H. I. & Tan, P. Gastric cancer pathology and underlying molecular mechanisms. Dig. Surg. 30, 150–158 (2013).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Busuttil, R. A. et al. An orthotopic mouse model of gastric cancer invasion and metastasis. Sci. Rep. 8, 825 (2018).
Furukawa, T., Kubota, T., Watanabe, M., Kitajima, M. & Hoffman, R. M. Orthotopic transplantation of histologically intact clinical specimens of stomach cancer to nude mice: correlation of metastatic sites in mouse and individual patient donors. Int. J. Cancer 53, 608–612 (1993).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Barker, N. et al. Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5+ve stem cells. Cold Spring Harb. Symp. Quant. Biol. 73, 351–356 (2008).
Leushacke, M. et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017).
Barker, N. et al. Lgr5+ve stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2, 540–552 (2012).
Ng, A. et al. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 16, 745–757 (2014).
Yamamoto, Y. et al. Overexpression of orphan G‐protein–coupled receptor, Gpr49, in human hepatocellular carcinomas with β‐catenin mutations. Hepatology 37, 528–533 (2003).
McClanahan, T. et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol. Ther. 5, 419–426 (2006).
Cao, H.-Z., Liu, X.-F., Yang, W.-T., Chen, Q. & Zheng, P.-S. LGR5 promotes cancer stem cell traits and chemoresistance in cervical cancer. Cell Death Dis. 8, e3039 (2017).
Sahin, U. et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin. Cancer Res. 14, 7624–7634 (2008).
Nemtsova, M. V. et al. Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci. Rep. 10, 504 (2020).
Hamilton, J. P. & Meltzer, S. J. A review of the genomics of gastric cancer. Clin. Gastroenterol. Hepatol. 4, 416–425 (2006).
Van der Flier, L. G. et al. The intestinal Wnt/TCF signature. Gastroenterology 132, 628–632 (2007).
Pek, M. et al. Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer. Oncogene 36, 4975–4986 (2017).
Park, J. W. et al. Multi-omics analysis identifies pathways and genes involved in diffuse-type gastric carcinogenesis induced by E-cadherin, p53, and Smad4 loss in mice. Mol. Carcinog. 57, 947–954 (2018).
Duan, S. et al. Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol. Med. Rep. 18, 3727–3736 (2018).
Xu, J. et al. Matrix metalloproteinase expression and molecular interaction network analysis in gastric cancer. Oncol. Lett. 12, 2403–2408 (2016).
Qiu, J., Sun, M., Wang, Y. & Chen, B. Identification of hub genes and pathways in gastric adenocarcinoma based on bioinformatics analysis. Med Sci. Monit. 26, e920261 (2020).
Jiang, B., Li, S., Jiang, Z. & Shao, P. Gastric cancer associated genes identified by an integrative analysis of gene expression data. BioMed. Res. Int. 2017, 7259097 (2017).
Chen, Y. et al. Identification of the collagen family as prognostic biomarkers and immune-associated targets in gastric cancer. Int. Immunopharmacol. 87, 106798 (2020).
Gao, X. et al. Alteration and prognostic values of collagen gene expression in patients with gastric cancer under different treatments. Pathol. Res. Pract. 216, 152831 (2020).
Li, Z. et al. Identifying multiple collagen gene family members as potential gastric cancer biomarkers using integrated bioinformatics analysis. PeerJ 8, e9123 (2020).
Zhang, Q.-N. et al. A panel of collagen genes are associated with prognosis of patients with gastric cancer and regulated by microRNA-29c-3p: an integrated bioinformatics analysis and experimental validation. Cancer Manag. Res. 11, 4757–4772 (2019).
Katoh, M., Kirikoshi, H., Terasaki, H. & Shiokawa, K. WNT2B2 mRNA, up-regulated in primary gastric cancer, is a positive regulator of the WNT–β-catenin–TCF signaling pathway. Biochem. Biophys. Res. Commun. 289, 1093–1098 (2001).
Zhang, Z., Wang, J. & Dong, X. Wnt2 contributes to the progression of gastric cancer by promoting cell migration and invasion. Oncol. Lett. 16, 2857–2864 (2018).
Rafi, J. H. et al. High expression of bone morphogenetic protein 1 (BMP1) is associated with a poor survival rate in human gastric cancer, a dataset approaches. Genomics 113, 1141–1154 (2021).
Liang, L. et al. Comprehensive evaluation of FKBP10 expression and its prognostic potential in gastric cancer. Oncol. Rep. 42, 615–628 (2019).
Wang, R.-G. et al. FKBP10 functioned as a cancer-promoting factor mediates cell proliferation, invasion, and migration via regulating PI3K signaling pathway in stomach adenocarcinoma. Kaohsiung J. Med. Sci. 36, 311–317 (2020).
Hirsch, D. et al. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis 35, 849–858 (2014).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Hagen, S. J. et al. Loss of tight junction protein claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology 155, 1852–1867 (2018).
Mankaney, G. et al. Gastric cancer in FAP: a concerning rise in incidence. Fam. Cancer 16, 371–376 (2017).
Bianchi, L. K. et al. Fundic gland polyp dysplasia is common in familial adenomatous polyposis. Clin. Gastroenterol. Hepatol. 6, 180–185 (2008).
Fang, D.-C. et al. Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J. Gastroenterol. 8, 787–791 (2002).
Sanz-Ortega, J. et al. LOH at the APC/MCC gene (5Q21) in gastric cancer and preneoplastic lesions: prognostic implications. Pathol. Res. Pract. 192, 1206–1210 (1996).
Wang, B. et al. LGR5 is a gastric cancer stem cell marker associated with stemness and the EMT signature genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS ONE 11, e0168904 (2016).
Wang, X. et al. LGR5 regulates gastric adenocarcinoma cell proliferation and invasion via activating Wnt signaling pathway. Oncogenesis 7, 57 (2018).
Wang, Z. & Liu, C. Lgr5-positive cells are cancer-stem-cell-like cells in gastric cancer. Cell. Physiol. Biochem. 36, 2447–2455 (2015).
Wu, C. et al. Lgr5 expression as stem cell marker in human gastric gland and its relatedness with other putative cancer stem cell markers. Gene 525, 18–25 (2013).
Yoon, J.-Y., Brezden-Masley, C. & Streutker, C. J. Lgr5 and stem/progenitor gene expression in gastric/gastroesophageal junction carcinoma—significance of potentially retained stemness. BMC Cancer 20, 860 (2020).
de Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Stange, D. E. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013).
Shultz, L. D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005).
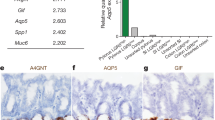
Tan, S. H. et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 578, 437–443 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Harrow, J. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7, S4.1–S4.9 (2006).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Durinck, S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Alexa A. & Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.42.0 (2020).
Acknowledgements
We would like to thank all members of the laboratory for technical expertise and insightful discussions. We would like to thank S. Mustafah and the staff at SIgN Flow Facility for assistance with FACS (SIgN, A*STAR, Singapore), L. Shuping, J. Lim and G. Wright (AMP, A*STAR, Singapore) for assistance with confocal imaging, and E. Cheah and staff at the SBIC-Nikon facility for assistance with bright-field imaging (A*STAR, Singapore). We also thank T. Ming, S. Srivastava and staff at the Department of Pathology, National University Hospital Singapore, for providing the human material, D. H. Alpers, (Washington University School of Medicine, USA) for providing the Gif-specific antibody, and W. Hunziker (IMCB, A*STAR, Singapore) for the ZO-1-specific antibody. Additionally, we would like to thank F. de Sauvage (Department of Molecular Biology, Genentech, USA) for providing the Lgr5-DTR–GFP mice. A.F. is supported by the National Medical Research Council (NMRC) Singapore under MOH-000366. N.B. is supported by A*STAR, the National Research Foundation (NRF; under NRFI2017-03) and the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant number 17H01399). Y.T. is supported by JSPS KAKENHI grant number 17H06710 and K.M. is supported by JSPS KAKENHI grant number 17K07161. The funding agencies had no role in the study design, data collection and analyses, decision to publish or preparation of manuscript.
Author information
Authors and Affiliations
Contributions
A.F. designed, performed all empirical experiments, collected and analysed data, and wrote the manuscript. Y.T. designed and performed OT and splenic injection experiments, collected and analysed data. S.S. performed all empirical experiments, collected and analysed data, and performed mouse husbandry. T.L.T. performed OT experiments. T.S. analysed human cancer data in the pathway analysis. S.H.T. provided advice and technical help with FACS and mouse cancer models. K.M. provided advice and technical help with human experiments and mouse cancer models. N.B. and Y.S. cloned and generated the Cldn18-IRES-CreERT2 mouse lines. N.A. analysed the RNA sequencing data and transcriptomics comparisons. R.R. performed pathological analyses on mouse tumours and quantified the immune cells. T.M. helped with the collection of patient samples. P.T. designed and supervised the cancer frequency analysis and helped with validation of the Cldn18 mouse model. B.L. designed, performed and supervised the analysis of RNA sequencing data, transcriptomics comparisons and gene expression analysis. N.B. designed and supervised the project, analysed the data and wrote the manuscript. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of a Cldn18–IRES–CreERT2 stomach–specific line.
(a) IF images of Cldn18 (red) and E–cadherin (green) co–staining in small intestine, colon, esophagus, liver and pancreas (n = 5 biological replicates). (b) IF images of tdTomato tracing at 24 hrs, 1 week and 6 months post–tamoxifen induction in the small intestine, colon, esophagus, liver and pancreas from Cldn18–dTom mice (n = 5 biological replicates). (c) IF images of Cldn18 (red) and E–cadherin (green) co–staining in pylorus regions from Cldn18–CreERT2 wild–type (top, n = 3 biological replicates), heterozygous (middle, n = 3 biological replicates) and homozygous (bottom, n = 3 biological replicates) mice. (d) IF images of Cldn18 (red) and E–cadherin (green) co–staining in corpus regions from Cldn18–CreERT2 wild–type (top, n = 3 biological replicates), heterozygous (middle, n = 3 biological replicates) and homozygous (bottom, n = 3 biological replicates) mice. (e) IF images of β–catenin (blue) and ZO–1 (magenta) co–staining in pylorus regions from Cldn18–CreERT2 wild–type (top, n = 3 biological replicates), heterozygous (middle, n = 3 biological replicates) and homozygous (bottom, n = 3 biological replicates) mice. (f) IF images of β–catenin (blue) and ZO–1 (magenta) co–staining in corpus regions from Cldn18–CreERT2 wild–type (top, n = 3 biological replicates), heterozygous (middle, n = 3 biological replicates) and homozygous (bottom, n = 3 biological replicates) mice. (g) IF images of pyloric lineage markers from uninduced Cldn18–dTom mice (top, n = 3 biological replicates) and induced mice (bottom, n = 3 biological replicates) 1 month post induction for Gastrin, Muc5 and Ki67 (yellow) and tdTomato (red). (h) IF images of corpus lineage markers from uninduced Cldn18–dTom mice (top, n = 3 biological replicates) and induced mice (bottom, n = 3 biological replicates) 1 month post induction for GIF, H+ pump, Muc5 and Ki67 (yellow) and tdTomato (red). Scale bars 100 μm.
Extended Data Fig. 2 Gastric tumour formation in various Cldn18-CreERT2-driven conditional mouse models.
H&E images of various allelic combinations tested. Tissue harvested for (1) Trp53fl/fl (1 yr, n = 15 biological replicates), (2) APCfl/WT + Trp53fl/fl (1 yr, n = 13 biological replicates), (3) KrasG12D (3 months, n = 8 biological replicates), (4) APCfl/fl (3 months, n = 9 biological replicates), (5) APCfl/fl + KrasG12D (1 month, n = 13 biological replicates), (6) Trp53fl/fl + KrasG12D (6 months, n = 4 biological replicates), (7) APCfl/fl + Trp53fl/fl (3 months, n = 22 biological replicates), (8) APCfl/fl + Trp53fl/fl + KrasG12D (2 months, n = 18 biological replicates) and (9) APCfl/WT + Trp53fl/fl + KrasG12D (4 months, n = 36 biological replicates) post–tamoxifen induction. Dotted lines demarcate the small intestine (SI), pylorus and corpus regions. Scale bars 1000 μm.
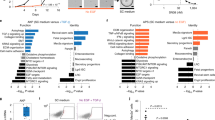
Extended Data Fig. 3 Comparison of human gastric cancer and Cldn18-ATK gastric tumour transcriptomes.
(a) PCA analysis for the Cldn18–ATK invasive tumour (green, n = 3 biological replicates) and neoplastic (purple, n = 3 biological replicates) samples. Strip–box plots visualizing the correlation between human normal (peach) or tumour (teal) samples with each Cldn18–ATK (b) invasive tumour and (c) neoplastic sample. The whiskers define the maxima and minima, the box is defined by the first and third quartile and the central line is the median for all data points.
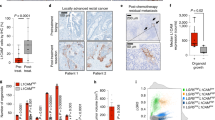
Extended Data Fig. 4 Cldn18–ATK mouse model accurately recapitulates advanced human gastric cancer.
(a) IHC images for β–catenin, E–cadherin, vimentin, parietal cell marker (proton (H+) pump) and F4/80 1 week post–tamoxifen (n = 5 biological replicates). (b) Representative H&E and IHC images for β–catenin, Trp53, Mapk (phosphorylated), Tff2, Muc5, E–cadherin, vimentin, GIF, Ki67, F4/80 and Cdx2 1 month post tamoxifen (n = 5 biological replicates). (c) IHC images for Mapk (phosphorylated), Trp53, Chief cell marker (GIF), Muc5 and Cdx2 at 2 months post–tamoxifen (n = 5 biological replicates). Invasive region is marked by blue line (d) IHC images for Mapk (phosphorylated), Trp53, Tff2 and F4/80 at the terminal stages of disease (n = 5 biological replicates). Blue lines demarcate primary tumour and invasive region. (E) Whole mount images of stomach tissue 1 week, 1 month and 2 month post–tamoxifen and lung metastasis highlighted with dotted black lines (n = 5 biological replicates). (f) H&E staining, IHC images for Mapk (phosphorylated), β–catenin, Trp53, Ki67, E–cadherin and vimentin for mouse liver metastases (n = 5 biological replicates). (g) IHC images for Mapk (phosphorylated), β–catenin and Trp53 in mouse lymph node metastases (n = 3 biological replicates). (h) H&E staining, IHC for Ki67, Cdx2 and Tff2 in mouse lung metastases (n = 5 biological replicates). Scale bars 500 μm.
Extended Data Fig. 5 Characterisation of Tumour Resident Lgr5+ stem–like cells.
Single channel IF images for GFP (green), along with (a) GIF – cyan and Muc5 – yellow, (b) H+ pump – red and Tff2 – blue and (c) Ki67 – magenta; for uninduced mice (control) and induced mice at 1 week, 1 month, 2 months, 3 months, terminal stages and invasive glands (n = 4, 5, 8, 5, 3 and 3 biological replicates respectively) in Cldn18–ATK tumours. Scale bars: 50 µm.
Extended Data Fig. 6 Tumour Resident Lgr5+ stem–like cells are required for disease progression.
(a) IHC images for Mapk, Cdx2, Tff2 and β–catenin in T–UT stomach (top), liver (middle) (n = 4 biological replicates) and T–DT treated stomach (bottom) (n = 9 biological replicates). (b) H&E and IHC images for GFP, Mapk, Cdx2, Ki67, Tff2 and β–catenin in DT treated liver. (c) IHC images for Mapk and β–catenin in DT treated and recovered stomach (top), magnified view of stomach and liver staining (n = 5 biological replicates). (d) IF staining for Ki67 (magenta, top) and Tff2 (yellow, bottom); IHC images for Mapk in T–UT (n = 4 biological replicates) and T–DT x10 stomach tissue (n = 3 biological replicates). (e) Quantification of changes in response to the three DT ablation strategies on tumour volumes. Statistical significance was determined by one way ANOVA and Tukey’s multiple comparisons test; ****adjusted P = < 0.0001. (f) Quantification of changes in proliferating cells after DT ablation. Statistical significance was determined by one way ANOVA and Tukey’s multiple comparisons test; **adjusted P = 0.0035. Graphs are presented as mean ± s.d. Scale bars: 100 µm Panel C macro: 500 μm.
Extended Data Fig. 7 Orthotopic transplantation of cancer organoids as a model to study gastric cancer.
(a) E–Cadherin, Cdx2 and Tff2 IHC images for uninduced gastric organoids and (b) gastric cancer organoids (n = 3 biological replicates). (c) Whole mount images and H&E images for stomach, liver and lung 12 weeks post orthotopic transplantation with Cldn18–dTom organoids. IHC images for tdTomato, GFP and Ki67 in the stomach (n = 5 biological replicates). (d) IHC images for GFP, tdTomato, Ki67, Mapk, β–catenin, E–Cadherin, Cdx2 and Tff2 in liver (top) and lung (bottom) metastases 12 weeks post orthotopic transplantation (n = 5 biological replicates). Scale bars: panels A and B 50μm, remaining panels 100 μm.
Extended Data Fig. 8 Tumour resident Lgr5+ stem-like cells are critical for the initiation and dissemination of gastric cancer in an orthotopic transplantation model.
(a) GFP IHC for stomach tissue (left) from mice receiving DT from 2, 6, 8, and 10 weeks post–transplantation. Liver metastases from mice receiving DT from 8 and 10 weeks post–transplantation and lung metastases from mice receiving DT at 10 weeks post transplantation (n = 5 biological replicates, per time point). (b) H&E staining (top panels) and GFP IHC for stomach tissue from untreated mice at 2, 6, 8 and 10 weeks post–transplantation. Liver and lung metastases at 8 and 10 weeks posttransplantation(n = 5 biological replicates, per time point). (c) GFP IHC for 12 week untreated stomach, liver and lung metastases (n = 5 biological replicates). Scale bars: 100 μm.
Extended Data Fig. 9 Characterising the Tumour Resident Lgr5+ stem–like cell regeneration.
(a) FACS gating for sorting GFP+ and GFP- epithelial cells (EpCam+) from untreated tumours (left), DT-treated tumours (middle) and wild-type unstained controls (right). (b) Relative expression of Lgr5, GFP, Axin2, Troy, Sox2 and Mist1 in the GFP– cells from normal vs tumour corpus epithelium (Normal epithelium: n = 3, tumour epithelium n = 6 biological replicate). Statistical significance was determined by unpaired t-test; ***P = 0.0004 (Lgr5), ***P = 0.0002 (GFP), ***P = 0.0005 (Sox2), ***P = 0.0001 (Mist1). (c) Relative expression of Axin2, Troy, Sox2 and Mist1 in the GFP– cells from untreated vs DT-treated tumour epithelium (untreated tumours n = 6, DT-treated tumour n = 14 biological replicates, except Mist1: DT-treated tumour n = 10 biological replicates). Statistical significance was determined by unpaired t-test; ****P = < 0.0001 (Troy), *P = 0.0116 (Sox2), **P = 0.0089 (Mist1). Graphs are presented as mean ± s.d.
Extended Data Fig. 10 Tumour Resident Lgr5+ stem-like cell ablation augments 5FU treatment.
(a) Timeline for 5FU and 5FU + DT treatment in tamoxifen–induced Cldn18–LATK mice. (b) Whole mount images for stomach and liver from untreated (left, n = 4 biological replicates), 5FU alone (middle, n = 9 biological replicates) and 5FU + DT treated mice (right, n = 13 biological replicates). Tumours and metastases are marked by dotted lines. (c) Percentage of mice displaying invasive tumours and distant metastases following treatment. (d) H&E (top), Ki67 (middle) and Lgr5 (bottom two) expression in untreated (left), 5FU alone (middle) and 5FU + DT treated mice (right). (e) Quantification of reduction in tumour volume following 5FU and 5FU + DT treatment. Statistical significance was determined by one way ANOVA and Tukey’s multiple comparisons test; ****adjusted P = < 0.0001 and *adjusted P = 0.0337. (f) Quantification of proliferating cells following 5FU and 5FU + DT treatment. Statistical significance was determined by one way ANOVA and Tukey’s multiple comparisons test; ****adjusted P = < 0.0001 and **adjusted P = 0.0073. Graphs are presented as mean ± s.d. Scale bars 500μm.
Supplementary information
Supplementary Tables
Supplementary Table 1: Summary of the Cldn18-CreERT2-driven conditional compound mutations generated, along with their respective sample size, latency period, tumour location and incidence of invasive and metastatic disease. Supplementary Table 2: Quantification of polymorphonuclear cells (H&E) and tumour-associated macrophages (F4/80 IHC) in the various Cldn18 cancer models. Supplementary Table 3: Quantification of polymorphonuclear cells (H&E) and tumour-associated macrophages (F4/80 IHC) at various time points in the Cldn18-ATK gastric tumours and associated metastases.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Fatehullah, A., Terakado, Y., Sagiraju, S. et al. A tumour-resident Lgr5+ stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat Cell Biol 23, 1299–1313 (2021). https://doi.org/10.1038/s41556-021-00793-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00793-9
This article is cited by
-
Somatic mouse models of gastric cancer reveal genotype-specific features of metastatic disease
Nature Cancer (2024)
-
POLQ inhibition attenuates the stemness and ferroptosis resistance in gastric cancer cells via downregulation of dihydroorotate dehydrogenase
Cell Death & Disease (2024)
-
Combined inhibition of Bcl-2 family members and YAP induces synthetic lethality in metastatic gastric cancer with RASA1 and NF2 deficiency
Molecular Cancer (2023)
-
Intracellular pH dynamics regulates intestinal stem cell lineage specification
Nature Communications (2023)
-
The composition and roles of gastric stem cells in epithelial homeostasis, regeneration, and tumorigenesis
Cellular Oncology (2023)