Abstract
Many animals have a conserved adaptive genome defence system known as the Piwi-interacting RNA (piRNA) pathway, which is essential for germ cell development and function. Disruption of individual mouse Piwi genes results in male but not female sterility, leading to the assumption that PIWI genes play little or no role in mammalian oocytes. Here, we report the generation of PIWI-defective golden hamsters, which have defects in the production of functional oocytes. The mechanisms involved vary among the hamster PIWI genes, whereby the lack of PIWIL1 has a major impact on gene expression, including hamster-specific young transposon de-silencing, whereas PIWIL3 deficiency has little impact on gene expression in oocytes, although DNA methylation was reduced to some extent in PIWIL3-deficient oocytes. Our findings serve as the foundation for developing useful models to study the piRNA pathway in mammalian oocytes, including humans.
Similar content being viewed by others
Main
Piwi-interacting RNAs (piRNAs) are derived from specific genomic loci termed piRNA clusters and form effector complexes with PIWI proteins, a germline-specific class of Argonaute proteins. These complexes guide recognition and silencing of their targets, which are mostly transposable elements (TEs)1,2,3. Mammalian PIWI–piRNA pathways have mostly been studied in mice, which express three PIWI proteins (PIWIL1 (also known as MIWI), PIWIL2 (also known as MILI) and PIWIL4 (also known as MIWI2)) abundantly in the testis but only weakly in the ovary1,2,3. These PIWI proteins bind distinct classes of piRNAs, which direct chromatin modifications of target TE sequences during embryogenesis and guide silencing of target TEs at the posttranscriptional level later in spermatogenesis, to ensure completion of meiosis and successful sperm production. A deficiency in Piwi genes in mice is characterized by spermatogenesis arrest and infertility in males, but females with deficiencies in these genes remain fertile4,5,6,7,8,9,10 with limited impact on TE silencing11,12. These findings clearly demonstrate that mouse Piwi genes are essential for male, but not female, germ cell development.
However, most mammalian species, including humans, possess an additional PIWI gene, termed PIWIL3, to the three PIWI genes described above13,14,15,16. PIWIL3 was found to be specifically expressed in oocytes and not in testes15,16,17. Thus, piRNA-mediated silencing may differ between mice and other mammals with four PIWI genes. However, little is known about the piRNA pathway in mammalian species with the four PIWI genes, particularly their potential roles in functional oocyte production. Golden Syrian hamsters (golden hamster, Mesocricetus auratus) have been used as an experimental rodent model for studying human diseases, particularly cancer and infectious diseases, including the recent Covid-19, because they display physiological and pharmacological responses resembling those of humans18. In addition, genome editing using the CRISPR–Cas9 system was recently enabled in golden hamsters to engineer the genes of interest19,20. Unlike laboratory mice and rats, which belong to the Muridae family of rodents lacking PIWIL3, the golden hamster belongs to the Cricetidae family of rodents and has four distinct PIWI genes. We recently found that hamster PIWIL1 and PIWIL2 are expressed in both the testis and the ovary, whereas PIWIL3 and PIWIL4 are exclusively expressed in the ovary and testis, respectively16.
In this article, we generate PIWI-deficient golden hamsters and address the question of whether PIWI genes are required for the production of functional oocytes. Our results identify the non-redundant essential role of PIWI genes in hamster oogenesis. Thus, this study implicates defects in PIWI genes in human infertility.
Results
Generation of PIWI-deficient golden hamsters using the CRISPR–Cas9 system
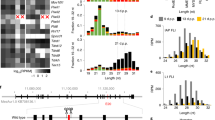
In the present study, we aimed to investigate the roles of PIWI genes in female reproduction by studying PIWIL1 and PIWIL3 genes, both of which are highly expressed in oocytes16. We directly injected Cas9 mRNA or Cas9 protein together with single guide RNAs (sgRNAs) into the pronucleus (PN) and/or cytoplasm of an embryo to generate hamsters deficient in PIWIL1 or PIWIL3. We injected the cells under a microscope with red filters (600 nm) in a dark room because even brief exposure of hamster zygotes to light results in total developmental arrest20. Injected embryos with normal morphology were transferred to each oviduct (10–15 embryos per oviduct) of pseudopregnant recipient females. In total, 46 pups were obtained from the PIWIL1-sgRNA-injected embryos, of which 23 carried a mutant allele, as demonstrated by genomic PCR and sequencing of tail tissue (Extended Data Fig. 1). However, nine female founder hamsters were infertile and could only establish three frameshift mutant hamster lines. For the production of PIWIL3-deficient hamsters, 18 pups were obtained from the injected embryos. Genomic sequencing revealed some mosaicism and identified a total of 13 types of PIWIL3 mutant alleles (Extended Data Fig. 2). We crossed F1 heterozygous mutant hamsters to generate F2 homozygous mutant hamsters, which yielded offspring with genotypes that segregated in a Mendelian distribution. Both PIWIL1-deficient hamsters and PIWIL3-deficient hamsters appeared normal without outwardly discernible morphological and behavioural abnormalities.
Abnormal spermatogenesis in PIWIL1-deficient golden hamsters
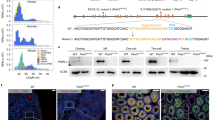
PIWIL1-deficient male hamsters displayed small testes and lack of mature sperm in the cauda epididymis (Extended Data Fig. 3a–e). Staining for DDX4 (mouse Vasa) and acrosome (with lectin peanut agglutinin (PNA)) revealed a complete lack of acrosome-positive S2 spermatids in the PIWIL1-deficient testes (Fig. 1a–d). In addition, the population of small RNAs of 29–31 nucleotides (nt) in length was markedly reduced in the PIWIL1-deficient testes (Fig. 1e–g). They showed a strong bias for uracil at their 5′ ends, which is a conserved characteristic of piRNAs1,2 (Fig. 1h). Genome mapping and annotation of testes PIWIL1–piRNAs revealed that they were mainly mapped to unannotated regions of the genome (Fig. 1i). These developmental abnormalities and reduction in corresponding piRNAs resembled those observed in Piwil1-deficient male mice3,4. PIWIL3 was not expressed in the testes16 and PIWIL3-deficient male hamsters displayed no overt phenotype.
a–d, Anti-DDX4 antibody (a–c) and PNA (d) staining of testis sections of 15-week-old wild-type and homozygous mutant (PIWIL1m/m) hamsters. Almost no spermatids after meiosis were observed in the PIWIL1-deficient testis. Enlarged views of the boxes in the lower panel of a show meiotic spermatocytes (arrow, b) and an early round spermatid (c). Similar results were observed in three independent samples. Scale bars, 100 µm (a and d) or 10 µm (b and c). e, Composition of small RNA categories according to their length distributions in testes from wild-type (PIWIL1w/w; left) and PIWIL1-deficient (right) golden hamsters. Small RNA reads from known ncRNAs (tRNA, rRNA, sn/snoRNA and miRNA) are indicated by the described colours. The reads were normalized to the reads mapped to miRNAs and the total number of obtained reads. f, An MA plot showing the expression level of each unique small RNA sequence. To calculate log2(fold change) and expression levels, 1 was added to each value to calculate the levels of small RNA sequences with 0 in either of the samples. Small RNAs decreased by more than eightfold in homozygous mutants were defined as PIWIL1–piRNA (indicated by blue lines). PIWIL1w/m, heterozygous mutant. g, The size distribution of PIWIL1–piRNA. Obtained small RNA reads were normalized to the total number of genome-mapped small RNA reads. h, WebLogo analysis of PIWIL1–piRNA. i, Annotation of genome-mapped PIWIL1–piRNA. Small RNA-seq was performed with three independent libraries and pooled data were used.
Defects in the production of functional oocytes in PIWI-deficient hamsters
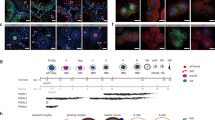
Western blots confirmed the lack of PIWIL1 and PIWIL3 in the mutant ovaries. Immunostaining also demonstrated the loss of PIWIL1 and PIWIL3 protein in the cytoplasm of mutant growing oocytes (Extended Data Fig. 4a,b). Histological examination of ovaries from PIWIL1-deficient and PIWIL3-deficient adult females revealed no gross abnormalities (Extended Data Fig. 4c). PIWIL1-deficient and PIWIL3-deficient female hamsters displayed complete sterility and reduced fertility, respectively (Fig. 2). When PIWIL1-deficient females were crossed with heterozygous males, they never became pregnant despite successful coitus, as demonstrated by the presence of sperm in the vagina (Fig. 2a,c). PIWIL3-deficient females when mated with heterozygous males displayed reduced fertility with both reduced pregnancy rates (43.5% versus 75% in homozygous (m/m) and heterozygous (w/m) females, respectively) and smaller litter sizes (4.6 versus 6.8 in homozygous and heterozygous females, respectively) (Fig. 2b,d). The homozygous PIWIL3-deficient pups proceeded into adulthood without expressing any overt developmental abnormalities.
a,b, The fecundity of female golden hamsters deficient in PIWIL1 (a) or PIWIL3 (b) was analysed by natural mating; n indicates the number of female hamsters used for each genotype. The numbers on the bar indicate pregnancy per coitus. *P = 0.0424 by two-sided Fisher’s exact test. c,d, Litter sizes from mated PIWIL1 mutant (c) and PIWIL3 mutant (d) females; n shows the number of mated females analysed and error bars indicate ±s.e.m. (**P = 0.0069 by two-sided t-test). e,f, The early embryogenesis ratio was analysed in vitro. 2C embryos (E1.5) from PIWIL1 heterozygous and homozygous mutant female hamsters mated with wild-type males were cultured for 1 day in vitro (e). 2C embryos (E1.5) from PIWIL3 heterozygous and homozygous mutant female hamsters mated with wild-type male were cultured for 1–2 days in vitro (f). (E2.5: ***P = 4.3 × 10–13, E3.5: ***P = 2.19 × 10–10 by χ2-test). M–B, morula and blastocyst. g–j, Representative images of E2.5 embryos derived from PIWIL1-deficient oocytes (g,h) and E3.5 embryos derived from PIWIL3-deficient oocytes (i,j). Embryos collected from three or four females were used, and similar results were confirmed more than three times. Note the arrested 3C and 5C embryos in the culture of PIWIL3-deficient embryos. Scale bars, 50 µm.
To examine how embryogenesis proceeds after fertilization with oocytes deficient in PIWIL1 or PIWIL3, we isolated two-cell (2C) embryos from PIWIL1-deficient and PIWIL3-deficient females crossed with wild-type males and cultured them in vitro. All maternal PIWIL1-deficient 2C embryos remained at the 2C stage and apparently died after 1 day of in vitro culture, whereas 2C embryos isolated from heterozygous females developed into four-cell (4C) to eight-cell (8C) embryos under the same culture conditions (Fig. 2e,g,h). Conversely, maternal PIWIL3-deficient embryos were significantly delayed in early embryogenesis, and 70% remained at the 2C or three-cell (3C) stage 1 day after in vitro culture (Fig. 2f). Even on the second day of culture, 30% remained at the 2C or 3C stage, and many others were arrested at the 4C or 8C stages including five-cell (5C) embryos, which indicates that one blastomere of the 2C was arrested and the other divided (Fig. 2f,i,j). These results indicate that both PIWIL1-deficient and PIWIL3-deficient oocytes can be fertilized and proceed through the first cell division, but they fail to develop, either completely or partially, at subsequent developmental stages. These results also highlight the non-redundant essential role of PIWI genes in hamster oogenesis.
Decrease in subclasses of piRNAs in PIWI-deficient oocytes
To gain insight into the molecular defects leading to the observed abnormalities, we performed small RNA sequencing (small RNA-seq) of PIWIL1-deficient and PIWIL3-deficient oocytes, and the results revealed a decrease in specific populations of small RNAs (Fig. 3a,c). In both heterozygous control oocytes, three peaks at 19 nt, 23 nt and 29 nt were observed. We compared the small RNA length distribution between heterozygous and homozygous oocytes by normalizing the entire small RNA population using the detected miRNA reads. This revealed a significant decrease in the population of 23- and 29-nt small RNAs in PIWIL1-deficient oocytes. In contrast, only the 19-nt small RNA population was decreased in PIWIL3-deficient oocytes. This is consistent with our recent findings that PIWIL1 binds to two populations of small RNAs, 23 and 29 nt (PIWIL1-bound piRNAs), while PIWIL3 binds to 19-nt small RNAs (PIWIL3-bound piRNAs) in metaphase II (MII) oocytes16. It has been recently shown that human PIWIL3 binds a class of ~20-nt small RNAs in oocytes17, which suggests that hamster PIWIL3 resembles the human orthologue.
a,c, Composition of small RNA categories according to their length distributions in hamster MII oocytes from PIWIL1 mutants (a) and PIWIL3 mutants (c). Small RNA reads from known ncRNAs (tRNA, rRNA, sn/snoRNA and miRNA) are indicated by the described colours. The reads were normalized to the reads mapped to miRNAs and the total number of obtained reads. b,d, MA plots showing the expression level of unique small RNA sequences in PIWIL1-deficient (b) and PIWIL3-deficient (d) oocytes. To calculate the log2(fold change) and expression levels, 1 was added to each value to calculate the levels of small RNA sequences with 0 in either of the samples. Small RNAs identical to the PIWIL1-bound piRNAs/PIWIL3-bound piRNAs and those that decreased by more than fourfold in homozygous mutants were defined as PIWIL1–piRNA or PIWIL3–piRNA, respectively (indicated by the blue lines). The red dots indicate piRNA bound to previously described PIWIs16. e,f, Size distributions of PIWIL1–piRNA (e) and PIWIL3–piRNA (f). Obtained small RNA reads were normalized to the total number of genome-mapped small RNA reads. g, WebLogo analysis of PIWIL1–piRNA (upper) and PIWIL3–piRNA (lower). h, Annotation of genome-mapped PIWIL1–piRNA (upper) and PIWIL3–piRNA (lower).
To further characterize the decreased population of small RNAs, changes in the expression level of each small RNA were analysed in PIWIL1 and PIWIL3 mutants. Decreased expression was observed in populations of small RNAs (Extended Data Fig. 5), which is consistent with our observation of the length distribution (Fig. 3a,c). Comparisons with the previously identified PIWIL1/PIWIL3-bound piRNAs16 indicated that the small RNAs, the populations of which were decreased, were identical to these PIWIL1/PIWIL3-bound piRNAs (Fig. 3b,d), which shows that the lack of PIWI proteins depleted piRNAs that bind to them. In contrast, the expression levels of a minor population of the PIWIL1/PIWIL3–piRNAs were not decreased in homozygous mutants. This may be because these piRNAs can be bound by other PIWI proteins and/or can be stably present in a PIWI-unassociated manner. Small RNAs identical to the PIWIL1-bound piRNAs/PIWIL3-bound piRNAs16 and those that decreased by more than fourfold in homozygous mutants were defined as PIWIL1–piRNA or PIWIL3–piRNA. As expected, PIWIL1–piRNAs were mostly 23- and 29-nt small RNAs, while PIWIL3–piRNAs were mostly 19-nt small RNAs (Fig. 3e,f). They both possessed uracil at their 5′ end (Fig. 3g). Genome mapping and annotation of PIWIL1–piRNAs and PIWIL3–piRNAs revealed that they were mainly mapped to unannotated regions of the genome, as in the case of mouse pachytene piRNAs3. A total of 12.4% PIWIL1–piRNAs and 15.5% of PIWIL3–piRNAs were mapped to the antisense orientation of TEs, which suggests that they can target TEs (Fig. 3h).
Altered expression of genes and TEs in PIWIL1-deficient oocytes
We also performed RNA-seq using samples isolated from PIWIL1-deficient and PIWIL3-deficient MII oocytes. This revealed that the lack of PIWIL1 significantly affected the oocyte transcriptome (Fig. 4a). We detected 1,612 differentially expressed genes (DEGs) in PIWIL1-deficient oocytes, 66.13% of which were upregulated, which indicates possible silencing of these genes by the PIWI–piRNA pathway. However, only 0.02% of the PIWIL1–piRNAs were mapped to the antisense direction of protein-coding genes (Fig. 3h). None of the upregulated DEGs were included in PIWIL1–piRNA-mapped protein-coding genes (PIWIL1–piRNA reads per kilobase of transcript per million mapped reads (RPKM) of >5), which suggests that PIWIL1–piRNAs cannot directly target mRNAs for their regulation. However, mouse PIWIL1 can bind spermatogenic mRNAs directly, without using piRNAs as guides, to form mRNP complexes that stabilize mRNAs essential for spermatogenesis21; therefore, hamster PIWIL1 may also regulate mRNAs via a piRNA-independent mechanism. In contrast, the level of most genes remained unchanged in PIWIL3-deficient oocytes, with only 21 DEGs detected (Fig. 4a). These results indicate that a number of genes are regulated by PIWIL1, but not by PIWIL3, in hamster oocytes.
a, MA plots showing the average expression level and log2(fold change) of the transcriptome in PIWIL1-deficient mutants (left) and PIWIL3-deficient mutants (right). Identified DEGs are indicated as red dots. b, Dot plots showing the expression levels of TEs in PIWIL1 mutants (left) and PIWIL3 mutants (right). Orange dots indicate TEs with more than twofold changes in the expression level. c, Examples of upregulated TE families in PIWIL1-deficient oocytes in three replicate (rep1–rep3) samples. Normalized bootstraps (n = 100) are plotted as boxplots for each of the three replicates. Each box spans from the first to the third quartile, and the bar inside the box shows the median. Whiskers above and below the box show 1.5 times the interquartile range.
The impact of the loss of PIWIL1 or PIWIL3 on the expression level of TEs was further analysed using RNA-seq. We recently re-sequenced the golden hamster genome and detected species-specific TEs16. Notably, ~80% of the expressed TEs (transcripts per kilobase million (TPM) of >1) were newly detected members of the TEs. An analysis of the TE expression levels revealed that PIWIL1 loss resulted in an increased expression of 29 families of TEs (by more than twofold) (Fig. 4b). In sharp contrast, the loss of PIWIL3 had little impact on the expression levels of TEs (Fig. 4b). In PIWIL1-deficient oocytes, TE family members, including ERV2-9, ERV2-14b and ERV2-14a, were upregulated the most (Fig. 4c). Of the 29 upregulated TE families, 18 were long terminal repeat (LTR) and 11 were long interspersed nuclear element (LINE) TEs. In addition, 27 out of the 29 upregulated TE families were specific to golden hamsters16, which suggests that PIWIL1 regulates active TEs that were recently added to the golden hamster genome. We then compared the abundance of the PIWIL1–piRNA population corresponding to each TE family with the change in expression of that TE in PIWIL1-deficient and PIWIL3-deficient oocytes (Fig. 5a,b). This revealed a correlation between PIWIL1–piRNA antisense abundance with particular TEs and the upregulation of these TEs with a lack of PIWIL1 but not a lack of PIWIL3. Together, these results show that PIWIL1 can regulate recently identified active TEs via their targeting by piRNAs in oocytes.
a,b, Differential expression levels between homozygous and heterozygous mutant samples for PIWIL1 (a) and PIWIL3 (b) are shown as the log2(fold change) along with piRNA levels. TE families with expression levels of >2 TPM in either homozygous or heterozygous mutants and >0 TPM in both mutants are listed. Increased expression levels are indicated in orange, whereas decreased expression levels are indicated in blue in the heatmaps. piRNA levels are shown as a bar graph, with the indicated small RNAs mapped to either the sense (grey) or antisense (red) directions of TEs. SINE, short interspersed nuclear element.
We performed gene ontology (GO) enrichment analysis of the DEGs identified with PIWIL1 deficiency and found enrichment of terms related to nucleosome assembly and transcriptional and epigenetic regulation (Extended Data Fig. 6a,b). This suggests that PIWIL1 loss may cause defects in chromatin and/or the genome integrity network. Consistent with this notion, PIWIL1-deficient 2C nuclei displayed a single enlarged nucleolus with altered nuclear DNA enrichment, while pronuclei derived from oocytes of all genotypes and 2C nuclei derived from PIWIL1 heterozygous and PIWIL3 homozygous oocytes displayed multiple nucleoli (Extended Data Fig. 7a,b). This suggests that PIWIL1 loss may induce the nucleolar stress response, which is often associated with cell cycle arrest and cell senescence22,23.
Altered DNA methylation in PIWIL3-deficient oocytes
In mice, nuclear Piwil4 acts as an effector for de novo DNA methylation of target TEs in embryonic male germ cells7,24,25,26. Recent studies have indicated that other PIWI proteins, which are predominantly cytoplasmic, may also function in DNA methylation in a Piwil4-independent manner in male germ cells27,28. These findings prompted us to examine DNA methylation in PIWIL1-deficient and PIWIL3-deficient oocytes, although both PIWIL1 and PIWIL3 are predominantly cytoplasmic in oocytes16 (Extended Data Fig. 4b). Because it is known that de novo DNA methylation is complete by the germinal vesicle (GV) stage in mouse oocytes29, we stained hamster PIWI-deficient and control GV oocytes with an anti-5-methylcytosine (5mC) antibody. The DNA methylation level was approximately the same in PIWIL1-deficient and control GV oocytes (Extended Data Figs. 8a and 9). However, PIWIL3-deficient oocytes had a significantly reduced DNA methylation level compared to control oocytes, as judged by 5mC staining (Extended Data Figs. 8b and 9). This suggests that PIWIL3 is involved in DNA methylation during oocyte development.
To explore the DNA methylation status in greater detail, we performed whole-genome bisulfite sequencing (Supplementary Table 1). This analysis revealed that the level of global CG methylation (DNA methylation at CG sites, which are the major methylation target) was significantly decreased in PIWIL3-deficient GV oocytes compared to that in heterozygous control oocytes (36.4% versus 39.1%, respectively) (Fig. 6a). The level of non-CG methylation was unchanged (Fig. 6b). We identified 4,364 differentially methylated regions (DMRs) using 3-kb windows, which included 3,581 less methylated DMRs and 683 more methylated DMRs (Fig. 6c). These DMRs were slightly more enriched in intergenic regions and tended to contain PIWIL3–piRNA target TEs (less methylated DMRs contained MT2C_Mm and AmnHarb1, whereas more methylated DMRs contained MER99, ORR1A3-int, MT2B1 and RMER1C) (Fig. 6d). However, the total CG methylation level at individual piRNA clusters, satellite repeats and TEs was not significantly altered in PIWIL3-deficient GV oocytes, except for four TEs, MT2C_Mm, MER136, MMTV-int and UCON58, among which MT2C_Mm was the only PIWIL3–piRNA target (Fig. 7a–c). While the overall CG methylation level of MT2C_Mm was reduced in PIWIL3-deficient oocytes (Fig. 7c), only one out of the total of 31 genomic copies of this TE was located in a less methylated DMR, which suggests that PIWIL3–piRNA binding to these TEs may not be the major cause of the observed DNA methylation changes. Our recent study16 showed that piRNA loading onto PIWIL3 is visible only in ovulated oocytes, which may preclude the probable piRNA-dependent function of PIWIL3 in intergenic DNA methylation in GV oocytes.
a,b, Violin plots showing the distributions of CG methylation (a) and non-CG methylation (b) levels of 3-kb windows (n = 415,096) determined by whole-genome bisulfite sequencing. Bisulfite sequencing was performed three times, each from independent biological replicates. The data shown represent pooled data from the three replicates. Crossbars indicate the median value. The global CG methylation levels are indicated above each graph. c, A scatterplot comparing the CG methylation levels of 3-kb windows in PIWIL3m/m and PIWIL3w/m GV oocytes. DMRs were defined as windows with a CG methylation difference of >20% and a P value of <0.05 (t-test). Significantly less methylated DMRs are plotted in blue, whereas more methylated DMRs are plotted in red. d, Genomic context of the DMRs. The DNA sequences of the windows from the whole genome (left), less methylated DMRs (middle) and more methylated DMRs (right) were used to analyse the presence of gene bodies, promoters, piRNA target TEs, non-piRNA target TEs, satellite repeats and unannotated intergenic regions.
a–c, Scatterplots describing the CG DNA methylation levels of individual piRNA clusters (a), satellite repeats (b) and TE families (c) in GV oocytes. PIWIL3–piRNA target TEs (TEs with more than 100 RPKM PIWIL3–piRNAs mapped) are plotted in red while the other transposons are plotted in grey. Names of differentially methylated TE families are indicated within the plots. d,e, Scatterplots describing the CG methylation levels in individual gene bodies (d) and gene promoters (e) in GV oocytes. Differentially methylated genes were defined as genes with >20% CG methylation differences between PIWIL3m/m and PIWIL3w/m, and P values between all heterozygous and homozygous replicates were <0.05. Among the differentially methylated genes, significantly less methylated genes were plotted in blue, whereas more methylated genes were plotted in red. P values were calculated by t-test. f, Boxplots showing the expression levels of genes with differential CG methylation found in their gene bodies or promoters. The left plot shows the expression levels of genes with more, less or unaltered methylation in gene bodies (n = 2,240, including 61 more methylated and 24 less methylated ones). The right plot shows the expression levels of genes with more, less or unaltered methylation in promoters (n = 10,465, including 35 more methylated and 20 less methylated ones). Each box spans from the first to the third quartile, and the bar inside the box shows the median. Whiskers above and below the box show 1.5 times the interquartile range. If the extreme data point is inside such a range, the whisker extends only to that point. P values were calculated using two-sided Mann–Whitney U-test. NS, not significant (P > 0.05); *P < 4.5 × 10–308. Exact P values below the threshold (4.5 × 10–308) are not shown due to software limitation.
Unlike any other differentiated mammalian cells, DNA methylation in oocytes occurs predominantly within gene bodies, with almost no methylation occurring in intergenic regions30. Therefore, we analysed the DNA methylation status of gene bodies and gene promoters in PIWIL3-deficient oocytes. We found that a number of genes showed altered levels of CG methylation at their gene bodies or promoters, with a larger population having reduced methylation, in PIWIL3-deficient GV oocytes (Fig. 7d,e). In addition, although there was no significant difference in oocyte transcripts between PIWIL3 mutant and controls (Fig. 4a), genes with increased DNA methylation at their gene bodies tended to be expressed at higher levels (Fig. 7f). These results show that PIWIL3 plays a role in DNA methylation in oocytes and suggest that the three-dimensional chromatin structure could be altered in PIWIL3-deficient oocytes. The significant decrease in 5mC staining intensity in PIWIL3-deficient GV oocytes (Extended Data Figs. 8 and 9) may also be explained by possible changes in antibody accessibility due to an altered chromatin structure. In addition, we found that meiosis occurred earlier in PIWIL3-deficient oocytes (Extended Data Fig. 10a), which suggests either premature entry into meiosis or accelerated post-ovulatory ageing. The morphology of the MII spindle was also altered in PIWIL3-deficient oocytes, with wider metaphase plates and longer acute-angled poles (Extended Data Fig. 10b). Although these phenotypes are modest (Extended Data Fig. 10c), they may suggest defects in chromosome segregation in PIWIL3-deficient oocytes and may account for the abnormal development of early embryos derived from PIWIL3-deficient oocytes.
Discussion
We analysed hamsters deficient in PIWIL1 or PIWIL3 and showed that maternally expressed PIWI genes are important for the development of preimplantation embryos. In support of this conclusion, Loubalova, Ogura, Svoboda and colleagues generated golden hamsters deficient in Mov10l1 (ref. 31), which is known to be an essential gene for the piRNA pathway in mice32,33. They demonstrated that both male and female Mov10l1-deficient hamsters are sterile and that piRNA production is markedly reduced in both the testes and the ovaries of Mov10l1-deficient hamsters. Sterility of female Mov10l1-deficient hamsters was found to be due to post-meiotic loss of developmental competence in zygotes. These phenotypes are reminiscent of those observed in the PIWIL1-deficient hamsters we produced. In addition, Zhang, Wu, Li and colleagues also produced golden hamsters deficient in piRNA pathway genes, including PIWIL1 and Mov10l1, and found that all female mutants are sterile34. Together, our results show the essential role of the piRNA pathway in the production of functional oocytes in golden hamsters.
PIWIL1 regulated the expression of genes, including TEs, while PIWIL3 had significantly different functions, such as DNA methylation with the lack of a strong effect on the expression of TEs and genes in oocytes. The origins of the piRNAs that bind to each PIWI are almost the same16, which suggests that the length of the piRNA, the PIWI–piRNA complex or the PIWI protein itself is responsible for its respective biological characteristics.
The establishment of DNA methylation in mouse oocytes and sperm is primarily due to the activity of DNMT3A and its cofactor DNMT3L35,36,37,38. DNMT3A-deficient and DNMT3L-deficient mouse oocytes can be fertilized normally, but embryos derived from these oocytes die by embryonic day 10.5 (E10.5), largely due to the absence of DNA methylation at maternally imprinted regions, which has led to the notion that the only definitive roles for oocyte DNA methylation are in post-fertilization contexts35,36,37. The observed DNA hypomethylation was modest, and transcripts from both TEs and protein-coding genes were not significantly affected in PIWIL3-deficient MII oocytes. How can cytoplasmic PIWIL3 (Extended Data Fig. 3b) be involved in DNA methylation, which is a nuclear event, without altering gene expression? It is worth noting that the mouse subcortical maternal complex, which plays an important role in DNA methylation and correct spindle formation in oocytes and pre-implantation embryos, is exclusively localized to the cytoplasm39. Defects in components of the subcortical maternal complex have been speculated to cause mislocalization of epigenetic factors such as DNMT proteins39. It will be interesting to examine the localization of factors involved in DNA methylation and/or spindle formation in PIWIL3-deficient oocytes. It is also tempting to speculate that PIWIL3 may be involved in the formation of developmentally competent chromatin, which could be inherited by the zygote, as is the case for DNMT3A-deficient and DNMT3L-deficient mice.
Although mice have contributed immensely to our understanding of the physiology of humans, it is also increasingly clear that the genetic and physiological differences between humans and mice hamper the extrapolation of the results obtained in mouse models to direct applications in humans. We infer that the emergence of an oocyte-specific Dicer isoform40 could reduce the impact of Piwi genes on the regulation of gene expression, thereby leading to the lack of the dependence on the piRNA pathway in the female germline of mice. The lack of discernible abnormalities in Piwi-deficient female mice may represent a special case of the piRNA pathway in mammals. Indeed, our study demonstrated that PIWI-deficient hamsters show abnormalities in oocyte function. This also suggests that PIWI genes should be studied as human infertility genes in women.
Methods
Animals
Golden hamsters were purchased from Japan SLC and maintained under 14-h light/10-h dark cycles. All animal experiments were approved by the Animal Care and Use Committee of Keio University and were conducted in compliance with the committee’s guidelines of Keio University.
Collection of GV oocytes, MII oocytes, PN zygotes and 2C embryos
Mature females were induced to superovulate via an intraperitoneal injection of 40 IU PMSG (ASKA Animal Health) between 12:00 and 14:00 on the day of conspicuous, post-oestrus vaginal discharge (day 1 of the oestrous cycle) for collecting GV and MII oocytes. GV oocytes were collected from the ovary at 102 h after PMSG injection. MII oocytes were collected from the oviduct at 115 h after PMSG injection. PN zygotes were collected from the oviduct after mating with males at the pro-oestrus stage. The 2C embryos were collected from the oviduct at 1 day after mating with males. Oocytes and embryos were collected from ovaries or oviducts with warmed and equilibrated HECM-10 medium41 and covered with paraffin oil (Nakarai). All embryos were then washed two-to-four times and cultured at 37.5 °C under 5% CO2. Culture dishes were pre-equilibrated for at least 5 h before use. The experiments were performed in a dark room with a >600-nm light.
Preparation of sgRNA and Cas9
The CRISPR–Cas9 target sites of PIWIL1 were designed using CRISPRdirect42 (http://crispr.dbcls.jp/) and produced using PCR-amplified templates and MEGA script T7 (Thermo Fisher Scientific). The PIWIL3 sgRNAs were designed using CRISPRdirect42 (http://crispr.dbcls.jp/) and synthesized using Alt-R CRISPR–Cas9 sgRNA (IDT) and annealed with Alt-R CRISPR–Cas9 tracrRNA (IDT). Cas9 was injected either as mRNA or protein. Cas9 mRNA was transcribed from the T7 promoter tagged Cas9 gene amplified from pX330 using a mMESSAGE mMACHINE T7 Transcription kit and a Poly(A) Tailing kit (Thermo Fisher Scientific). Cas9 protein was purchased from IDT. All primers are listed in Supplementary Table 2.
Generation of PIWIL1-deficient and PIWIL3-deficient golden hamsters
The technique for PN microinjection in golden hamsters is the same as that for mice, but with embryo manipulation conditions specifically developed for hamsters. In brief, fully equilibrated HECM-10 medium (37.5 °C, 5% CO2) covered with mineral oil was used as the injection medium. Embryos from one female were transferred to a 10 µl HECM-10 drop on a microinjection dish and 600 nM CRISPR RNA and 200 nM Cas9 mRNA (PIWIL1) or chemically synthesized 1 µM sgRNA and 1 µM Cas9 protein complex (PIWIL3) solution was injected into the PN and/or cytoplasm of an embryo. Microinjections were performed on a heated microinjection stage (37 °C) with red filters (cut-off < 600 nm), and all embryo handling procedures were performed in a dark room. After injection, embryos were transferred to the oviducts of pseudopregnant females within 30 min. Recipients were allowed to naturally deliver and raise their pups.
Establishment of PIWIL1-deficient and PIWIL3-deficient lines
Founder pups were mated with wild-type males or females to produce the F1 generation. Genomic DNA was extracted from small pieces of tail tissue from F1 pups. Approximately 1-kb genomic fragments containing the target site were amplified by PCR using specific primers, and direct sequencing was performed. All primers are listed in Supplementary Table 2. The PIWIL1-deficient mutant line (15Adel4) and the PIWIL3-deficient mutant line (26Adel192) used in the present study were mated at least five times with wild types to avoid off-target effects. PIWI-deficient golden hamster lines were maintained by mating with a wild type purchased from a breeder once every 6 months because golden hamsters are a closed colony.
Immunofluorescence
Ovaries from wild-type and mutant female golden hamsters were fixed with 4% paraformaldehyde for preparing paraffin-embedded sections. Paraffin-embedded tissue blocks were cut into 3-µm slices, heated with antigen retrieval buffer (100× Tris-EDTA buffer, pH 9.0; Abcam) in a microwave for 2 min after boiling and treated with anti-PIWIL1 monoclonal antibody (1A5)13 or anti-PIWIL3 monoclonal antibody (3E12)16. An Alexa-488-conjugated goat anti-mouse IgG (Thermo Fisher) was used as the secondary antibody. For staining the acrosome, 3-µm paraffin-embedded testis sections were treated with PNA–FITC (Sigma-Aldrich). To visualize genomic DNA, the sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI; Dojindo). Fluorescence was observed using an IX71 fluorescence microscope (Olympus).
Western blotting
Lysates of the MII oocytes were prepared by direct lysis with 2× SDS protein sample buffer. Total protein from 20 MII oocytes was separated on SDS–polyacrylamide gels and transferred to polyvinylidenedifluoride membranes. PIWIL1, PIWIL3 and TUBB were visualized using an ECL detection system (GE Healthcare) after incubation with anti-PIWIL1 monoclonal antibody (1A5), anti-PIWIL3 monoclonal antibody (3E12) or anti-TUBB monoclonal antibody (DSHB, E7), respectively. Horseradish-peroxidase-linked sheep anti-mouse IgG (MP Biomedicals) was used as the secondary antibody.
Fertility analysis
Mutant female golden hamsters at the proestrus stage (2–4 months old) were caged overnight with males at a 1:1 ratio, and vaginal sperm was examined the following morning. The pregnancy rate was calculated according to the number of successful deliveries per observation of sperm in the vagina.
Small RNA-seq
Total RNA was extracted from MII oocytes of PIWIL1 and PIWIL3 homozygous and heterozygous mutants and from testes of 7-week-old PIWIL1 homozygous mutants and wild types for small RNA-seq. The construction of small RNA libraries was performed using a NEXTFLEX Small RNA-seq kit v.3 (PerkinElmer) according to the manufacturer’s instructions. The smallRNA libraries were sequenced using the MiSeq (Illumina) platform, with three replicates from different samples sequenced for each mutant.
Small RNA-seq data analysis
Small RNA-seq analysis was performed as previously described43 with slight modifications. Briefly, the adapter sequence and 4-nt random sequences at the 3′ and 5′ ends were removed using cutadapt (v.1.14)44. A quality filter of 30 was adopted by filtering sequences below this threshold. Reads in the range of 10–40 nt after adapter removal were used for further analysis. Reads were mapped to the hamster reference genome (hamster.sequel.draft-20200302.arrow.fasta)16 using Bowtie (v.1.1.1)45, permitting 0 mismatches, and genome-mapped reads were used for further analysis. Genome-mapped reads were compared between replicate samples (n = 3 per sample) and combined because of their high reproducibility (r2 = 0.71~0.95). Reads were further mapped to known transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear/small nucleolar RNA (sn/snoRNA) and microRNA (miRNA) sequences. The tRNA sequences were obtained from RepeatMasker, while rRNA, sn/snoRNA and miRNA sequences were obtained from Ensemble BioMart46. The read count mapped to miRNAs was used to normalize small RNA reads obtained from heterozygous and homozygous mutants. Read counts mapped to each unique small RNA sequence were compared between heterozygous and homozygous samples, and small RNAs identical to the PIWIL1-bound piRNAs/PIWIL3-bound piRNAs16 and those that decreased by more than fourfold in homozygous mutants were defined as PIWIL1–piRNAs or PIWIL3–piRNAs, respectively. The reads mapped to known noncoding RNAs (ncRNAs) were removed from the PIWIL1–piRNAs and PIWIL3–piRNAs. Nucleotide composition was analysed using WebLogo (v.3.7.5)47, and annotation of the PIWIL1–piRNAs and PIWIL3–piRNAs were assigned using a previously described script48. The priority of the assignment was in the order of transposons, repeats and protein-coding genes. The reads were assigned to the highest priority annotation; therefore, every read was assigned to a single annotation. PIWIL1–piRNAs and PIWIL3–piRNAs were further mapped to the consensus sequence of transposon families obtained from the RepeatMasker database and our previous research16. The mapping to transposon consensus sequences was performed using Bowtie (v.1.1.1)45, with up to two mismatches permitted. Mapped reads were counted and normalized to the total number of mapped reads and the length of each transposon sequence. For small RNA-seq samples from testes, analysis was performed as described for MII oocyte samples except for the definition of PIWIL1–piRNAs. Read counts mapped to each unique small RNA sequence were compared between wild-type and homozygous mutant samples, and small RNAs that decreased by more than eightfold in homozygous mutants were defined as PIWIL1–piRNAs in the testis.
RNA-seq of MII oocytes
Total RNA was purified from 50 MII oocytes of 2–4-month-old golden hamsters using a NucleoSpin RNA Plus XS kit (Macherey-Nagel) for RNA-seq. RNA-seq libraries were constructed using total RNA from MII oocytes and a SMART-Seq Stranded kit (TaKaRa Bio). Sequencing was performed using the NovaSeq 6000 (Illumina) platforms.
RNA-seq data analysis
Prepared libraries were sequenced in paired ends, and triplicate samples were obtained per experiment. Low-quality sequences (quality value lower than 30) and 3-nt random sequences were removed using cutadapt (v.1.14)44 software. Mapping to the golden hamster genome (hamster.sequel.draft-20200302.arrow.fasta)16 was performed using HISAT (2.2.0)49 with default parameters. featureCount (v.2.01)50 was used to count reads mapped to genes. featureCounts output was further processed using TCC-GUI (v.2019.02.08)51 with default parameters. TCC-GUI uses the TMM normalization method, and edgeR was used to filter DEGs. Extracted DEGs were converted to mouse homologue genes using Ensemble BioMart46, and DAVID52 was used to perform GO enrichment analysis. Salmon (v.1.3.0)53 and Sleuth (v.0.30.0)54 were used for quantification of the expression levels of TEs. Wasabi (v.1.0.1) was used to analyse the Salmon output in Sleuth. To map the TE reads, consensus sequences of TEs were obtained from RepeatMasker and our previous analysis16, and the TE sequences were merged with the other gene sequences. The mean value for three replicates was used to determine the overall expression levels of TEs.
MII spindle staining
MII oocytes were freed from the zona pellucida by treatment with acidic Tyrode’s solution. Zona-free GV oocytes were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X‐100 in 0.01% PVA/PBS for 2 min at room temperature. For immunofluorescence staining, we used Alexa-488-conjugated tubulin-α monoclonal antibody (MBL). Cells were incubated at 4 °C overnight with a primary antibody and then washed three times. To visualize genomic DNA, the cells were counterstained with DAPI (Dojindo). The spindle fluorescence images were captured using a FV3000 laser scanning confocal microscope (Olympus). The z-stack images were reconstructed, and spindle lengths were measured using Imaris software (Zeiss).
5mC staining
GV oocytes were freed from the zona pellucida by treatment with acidic Tyrode’s solution.
Zona-free GV oocytes were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X‐100 in 0.01% PVA/PBS for 2 min at room temperature. These cells were treated with 2 N HCl 10 min at 37 °C and washed twice with 0.01% PVA/PBS. For immunofluorescence staining, we used a monoclonal primary antibody against 5mC (33D3, Abcam). Cells were incubated at 4 °C overnight with the primary antibody and then washed and incubated with an Alexa-Fluor-488-conjugated anti-mouse IgG secondary antibody (Thermo Fisher Scientific) at room temperature for 1 h. Fluorescence was measured using an IX71 fluorescence microscope (Olympus).
Whole-genome bisulfite sequencing and data analysis
DNA from 30 GV oocytes was spiked with 1% unmethylated lambda phage DNA. Libraries were generated using the post-bisulfite adaptor tagging method with eight cycles of library amplification55,56 and sequenced using the NovaSeq 6000 platform (NVCS v.1.6 and RTA v.3.4.4)57. Data analysis was performed as previously described56. Briefly, reads were trimmed to remove low-quality bases and were mapped to the golden hamster genome using Bismark (v.0.20.0). Methylation data at CG and non-CG sites covered with 5–500 reads were extracted and compared between the biological triplicates in 3-kb windows. After confirmation of the reproducibility (Pearson’s correlation coefficient >0.84 for PIWIL3w/m and >0.92 for PIWIL3m/m), the triplicate data were combined and used for downstream DMR analyses. DMRs were defined as windows with a CG methylation difference of >20% and a P value of <0.05 (t-test).
Statistics and reproducibility
Statistical significance was determined using two-sided Fisher’s exact test (Fig. 2b), two-sided t-test (Figs. 2d and 6c and Extended Data Figs. 8 and 10), χ2-test (Fig. 2f) or two-sided Mann–Whitney U-test (Fig. 7f) as described in the figure legends. Immunostaining (Fig. 1a–d and Extended Data Figs. 3e, 4b, 8a,b9 and 10) and observation of embryo morphology (Fig. 2g–j and Extended Data Fig. 7) were performed independently at least three times. Three independent RNA-seq (Figs. 1e–i and 3–5 and Extended Data Figs. 5 and 6) and whole-genome bisulfite sequencing (Fig. 6) libraries were constructed from three independent pooled embryo samples. No statistical method was used to predetermine sample sizes. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The raw sequence data generated during this study are available from the Gene Expression Omnibus repository under accession number GSE164356. Requests for materials should be addressed to H. Siomi. Source data are provided with this paper. All data presented in this study are available in the main text or the supplementary materials.
References
Iwasaki, Y. W., Siomi, M. C. & Siomi, H. PIWI-Interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019).
Pillai, R. S. & Chuma, S. piRNAs and their involvement in male germline development in mice. Dev. Growth Differ. 54, 78–92 (2012).
Deng, W. & Lin, H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002).
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Kuramochi-Miyagawa, S. et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887–892 (2010).
Reuter, M. et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011).
De Fazio, S. et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011).
Cheng, E. C., Kang, D., Wang, Z. & Lin, H. PIWI proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells. PLoS ONE 9, e97821 (2014).
Lim, A. K. et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development 140, 3819–3825 (2013).
Kabayama, Y. et al. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 45, 5387–5398 (2017).
Hirano, T. et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20, 1223–1237 (2014).
Sasaki, T., Shiohama, A., Minoshima, S. & Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 82, 323–330 (2003).
Roovers, E. F. et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 10, 2069–2082 (2015).
Ishino, K. et al. Hamster PIWI proteins bind to piRNAs with stage-specific size variations during oocyte maturation. Nucleic Acids Res. 49, 2700–2720 (2021).
Yang, Q. et al. Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat. Commun. 10, 3389 (2019).
Sia, S. F. et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020).
Hirose, M. et al. Acrosin is essential for sperm penetration through the zona pellucida in hamsters. Proc. Natl Acad. Sci. USA 117, 2513–2518 (2020).
Fan, Z. et al. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS ONE 9, e109755 (2014).
Vourekas, A. et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 19, 773–781 (2012).
Korsholm, L. M. et al. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 48, 9449–9461 (2020).
Tanaka, H. et al. The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Rep. 18, 2148–2161 (2017).
Molaro, A. et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 28, 1544–1549 (2014).
Zoch, A. et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 584, 635–639 (2020).
Watanabe, T., Cui, X., Yuan, Z., Qi, H. & Lin, H. MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. 37, e95329 (2018).
Nagamori, I. et al. Comprehensive DNA methylation analysis of retrotransposons in male germ cells. Cell Rep. 12, 1541–1547 (2015).
Yang, F. et al. TEX15 associates with MILI and silences transposable elements in male germ cells. Genes Dev. 34, 745–750 (2020).
Sasaki, H. & Matsui, Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140 (2008).
Demond, H. & Kelsey, G. The enigma of DNA methylation in the mammalian oocyte. F1000 Res. https://doi.org/10.12688/f1000research.21513.1 (2020).
Loubalova, Z. et al Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00746-2 (2021).
Zheng, K. et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Natl Acad. Sci. USA 107, 11841–11846 (2010).
Frost, R. J. A. et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl Acad. Sci. USA 107, 11847–11852 (2010).
Zhang, H. et al. The piRNA pathway is essential for generating functional oocytes in golden hamsters. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00750-6 (2021).
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. & Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823 (2004).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Smallwood, S. A. et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 (2011).
Monk, D., Sanchez-Delgado, M. & Fisher, R. NLRPs, the subcortical maternal complex and genomic imprinting. Reproduction 154, R161–R170 (2017).
Flemr, M. et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155, 807–816 (2013).
Ludwig, T. E., Squirrell, J. M., Palmenberg, A. C. & Bavister, B. D. Relationship between development, metabolism, and mitochondrial organization in 2-cell hamster embryos in the presence of low levels of phosphate. Biol. Reprod. 65, 1648–1654 (2001).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Iwasaki, Y. W. et al. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol. Cell 63, 408–419 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Kinsella, R. J. et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011, bar030 (2011).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Iwasaki, Y. W., Ishino, K. & Siomi, H. Deep sequencing and high-throughput analysis of PIWI-associated small RNAs. Methods 126, 66–75 (2017).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Su, W., Sun, J., Shimizu, K. & Kadota, K. TCC-GUI: a Shiny-based application for differential expression analysis of RNA-seq count data. BMC Res. Notes 12, 133 (2019).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Pimentel, H., Bray, N. L., Puente, S., Melsted, P. & Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Au Yeung, W. K. et al. Histone H3K9 methyltransferase G9a in oocytes Is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep. 27, 282–293 e284 (2019).
Toh, H. et al. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics 18, 31 (2017).
Acknowledgements
We thank all members of the Siomi Laboratory, especially H. Ishizu and K. Murano, for discussions and comments on this work, I. Ishimatsu for preparing paraffin sections, and H. Nishihara (Tokyo Institute of Technology) for his comments on hamster TEs. We also thank J. Oishi of the Sasaki Laboratory and T. Akinaga (Laboratory for Research Support, Medical Institute of Bioregulation, Kyushu University) for their help with whole-genome bisulfite sequencing. We are grateful to P. Svoboda (IMG, Czech Academy of Sciences), J. Li (Model Animal Research Center, Nanjing Medical University) and L. Wu (Shanghai Institute of Biochemistry and cell Biology, CAS) for sharing unpublished data. We are also grateful to M. Okabe and M. Ikawa (Osaka University) for their comments on the manuscript. Funding included support from a Grant-in-Aid for the Japan Society for the Promotion of Science (JSPS) KAKENHI grant no. 20H03175 (to H.H.), the Takeda Science Foundation (to H.H.), a JSPS Fellows (18J22025) (to K.I.), from JSPS KAKENHI grant nos 19H05268 and 18H02421, and from JST PRESTO grant no. JPMJPR20E2 (to Y.W.I.), for Scientific Research (S) (25221003) and the Program of totipotency (19H05753) (to H. Siomi). H. Siomi is also a recipient of funding for the project for elucidating and controlling mechanisms of ageing and longevity (1005442) from the Japan Agency for Medical Research and Development (AMED). This work was also supported by JSPS KAKENHI grant no. JP18H05214 (H. Sasaki).
Author information
Authors and Affiliations
Contributions
H. Siomi and H.H. designed the experiments. H.H. generated the PIWIL1-deficient and PIWIL3-deficient hamsters (with the help of H.M.). K.I. and Y.W.I. conducted small RNA-seq and RNA-seq and analysed the obtained data. W.K.A.Y. and H. Sasaki conducted whole-genome bisulfite sequencing and analysed the obtained data. H. Siomi, H.H., Y.W.I. and H. Sasaki wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks René Ketting, Jeremy Wang and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Strategy for the production of PIWIL1-deficient golden hamsters by CRISPR/Cas9.
a, Structure of the golden hamster PIWIL1 gene and target locus for sgRNA. b, The guide RNA sequence for the golden hamster PIWIL1 gene. c, Production of PIWIL1-deficient golden hamster lines. d, The PIWIL1 nucleic acid and amino acid sequences of the targeted site in wild-type hamsters. Upper case underlined denotes the exon sequence, bold with a red box denotes the target sequence, and red denotes the deleted sequence in the 15Adel4 line. e, The PIWIL1 nucleic acid and amino acid sequence of the targeted site in the mutant line (15 A del4). The amino acid sequences in red indicate sequences changed by deletion.
Extended Data Fig. 2 Strategy for the production of PIWIL3-deficient golden hamster by CRISPR/Cas9.
a, Structure of the golden hamster PIWIL3 gene and target loci for sgRNAs. b, The guide RNA sequences for the golden hamster PIWIL3 gene. c, Production of PIWIL3-deficient golden hamster lines. d, The PIWIL3 nucleic acid and amino acid sequence of targeted site in wild-type hamsters. Upper case underlined denotes the exon sequences, bold with a red box denotes the target sequences, and red denotes the deleted sequence in the 26Adel192 line. e, The PIWIL1 nucleic acid and amino acid sequence of targeted site in the mutant line (26 A del192). The amino acid sequences in red indicate sequences changed by deletion.
Extended Data Fig. 3 Defects in spermatogenesis in PIWIL1-deficient male golden hamsters.
a, Testis and cauda epididymis of 1-week-old (1w), 4-week-old (4w) and 10-week-old (10w) from wild-type and PIWIL1-deficient golden hamsters. Scale bars = 10 mm. b, Sperm from the 10w wild-type and PIWIL1-deficient cauda epididymis. Scale bars = 50 µm. c, d, Hematoxylin and eosin-stained 4w (c) and 15w (d) testis sections of wild-type and PIWIL1m/m hamsters, respectively. Scale bars = 100 µm. e, anti-PIWIL1 antibody staining of testis sections of 15-w wild-type and PIWIL1m/m hamsters. Scale bars = 100 µm.
Extended Data Fig. 4 Lack of PIWI proteins in oocytes of PIWIL1- or PIWIL3-deficient golden hamsters.
a, Total lysates of 20 MII oocytes each from heterozygous mutant (w/m) and homozygous mutant (m/m) golden hamsters were subjected to western blotting. TUBB was used as the loading control. Spectra™ Multicolor Broad Range Protein Ladder (Thermo Fisher Scientific) was used as size marker. b, Immunological staining of PIWIL1- and PIWIL3-deficient ovaries with an anti‐PIWIL1 and PIWIL3 antibodies. The arrowheads in the mutants indicate the ovarian follicles. The zona pellucida fixed with paraformaldehyde showed autofluorescence. In particular, PIWIL1, which requires a long exposure time, has strong ring-shaped fluorescence both in the wild type and PIWIL1m/m. Scale bars = 50 µm. c, Ovarian sections from 8‐week‐old wild-type and PIWIL1- and PIWIL3-deficient golden hamsters. Hematoxylin and eosin‐stained ovarian sections of wild-type, PIWIL1- and PIWIL3-deficient golden hamsters. Scale bars = 200 µm. Note: By fixing PIWIs, the antigenicity is weakened. PIWIL1 antigenicity is particularly weak even after antigen retrieval treatment: therefore, the exposure time is long and the background of the zona pellucida becomes visible.
Extended Data Fig. 5 Changes in the expression levels of each unique small RNA sequence in PIWI-deficient mutants.
An MA plot showing the expression level of each unique small RNA sequence. To calculate log2 fold change and expression levels, 1 was added to each value to calculate the levels of small RNA sequences with 0 in either sample.
Extended Data Fig. 6 Gene ontology analysis of MII oocytes in PIWIL1-deficient golden hamsters.
a, b, Enriched gene ontology (biological process) terms using DEGs obtained in PIWIL1-deficient MII oocyte transcriptome analysis. Upregulated genes in the PIWIL1-deficient MII oocytes (a). Downregulated genes in the PIWIL1-deficient MII oocytes (b). Terms with p-values lower than 0.001 are listed.
Extended Data Fig. 7 Nucleolus abnormalities in 2 C embryos derived from PIWIL1-deficient oocytes.
a, Pro-nucleus morphology of the zygotes. There were no pro-nucleus morphology differences in maternally PIWIL1- and PIWIL3-deficient zygotes. b, Nucleus morphology of the 2 C embryos. PIWIL1m/m showed a single large nucleolus, while PIWIL1w/m and PIWIL3m/m formed multiple small nucleoli. Enlarged view of box showed nucleus and arrows indicate the nucleolus. Scale bars = 50 µm.
Extended Data Fig. 8 DNA methylation in PIWIL1- and PIWIL3-deficient GV oocyte.
a, b, 5mC staining of PIWIL1-deficient (a) and PIWIL3-deficient GV oocytes (b). The amounts of 5mC were determined by measuring the fluorescence intensity using image J software (right). n indicates the number of independent GV oocytes collected from three females and used for analysis (Extended Data Fig. 9), and error bars indicate ± s.d. (***P=4.01e-8 by 2-sided t-test) Scale bars = 50 µm.
Extended Data Fig. 9 Representative GV oocytes stained with anti-5mC antibody.
GV oocytes were stained with a 5mC antibody (33D3) and the fluorescence GV nuclei were imaged. The fluorescence was analyzed by ImageJ and used to be described in Extended Data Fig. 8. Scale bars = 25 µm.
Extended Data Fig. 10 Meiosis and MII spindle formation in ovulated oocytes of PIWIL3-deficient golden hamsters.
a, PIWIL3w/m and PIWIL3m/m oocytes were collected, and the stages of meiosis at 7 and 10 am in the morning were analyzed following TUBA and DAPI staining. b, MII oocytes at 10 am in the morning were stained with anti-TUBA antibody (green) and DAPI (blue). Scale bars = 5 µm. c, The size of the MII spindles (A and B in Extended Data Fig. 10b) was measured and compared between PIWL3w/m and PIWL3m/m. n indicates the number of independent MII oocytes collected from three females and used for analysis, error bars indicate ± s.d. (A: ***P=0.000417, B: *P=0.0377 by 2-sided t-test).
Supplementary information
Supplementary Tables
Supplementary Table 1: Sequencing and mapping summary of whole-genome bisulfate sequencing. Supplementary Table 2: Primers and gRNA.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 4
Gel image.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Hasuwa, H., Iwasaki, Y.W., Au Yeung, W.K. et al. Production of functional oocytes requires maternally expressed PIWI genes and piRNAs in golden hamsters. Nat Cell Biol 23, 1002–1012 (2021). https://doi.org/10.1038/s41556-021-00745-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00745-3
This article is cited by
-
Divergent composition and transposon-silencing activity of small RNAs in mammalian oocytes
Genome Biology (2024)
-
Themes and variations on piRNA-guided transposon control
Mobile DNA (2023)
-
The non-redundant functions of PIWI family proteins in gametogenesis in golden hamsters
Nature Communications (2023)
-
Emerging roles and functional mechanisms of PIWI-interacting RNAs
Nature Reviews Molecular Cell Biology (2023)
-
Mammalian genome innovation through transposon domestication
Nature Cell Biology (2022)