Abstract
Heterochromatin, typically marked by histone H3 trimethylation at lysine 9 (H3K9me3) or lysine 27 (H3K27me3), represses different protein-coding genes in different cells, as well as repetitive elements. The basis for locus specificity is unclear. Previously, we identified 172 proteins that are embedded in sonication-resistant heterochromatin (srHC) harbouring H3K9me3. Here, we investigate in humans how 97 of the H3K9me3-srHC proteins repress heterochromatic genes. We reveal four groups of srHC proteins that each repress many common genes and repeat elements. Two groups repress H3K9me3-embedded genes with different extents of flanking srHC, one group is specific for srHC genes with H3K9me3 and H3K27me3, and one group is specific for genes with srHC as the primary feature. We find that the enhancer of rudimentary homologue (ERH) is conserved from Schizosaccharomyces pombe in repressing meiotic genes and, in humans, now represses other lineage-specific genes and repeat elements. The study greatly expands our understanding of H3K9me3-based gene repression in vertebrates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq, ChIP–seq and srHC-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE154233. Previously published sequencing data that were reanalysed here are available from GEO under accession codes GSE87039, GSM817237, GSM817240, GSM958154, GSM521917, GSM521915 and GSM1127120, or from the 4D nucleome under accession code 4DNFID41C3X7. Source data are provided with this paper. All other data supporting the finding of this study are available from the corresponding author on reasonable request.
Code availability
All code for data analysis and visualization is available from the corresponding author on request.
Change history
29 September 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41556-021-00759-x
References
Becker, J. S., Nicetto, D. & Zaret, K. S. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29–41 (2016).
Becker, J. S. et al. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell 68, 1023–1037 (2017).
Margueron, R. & Reinberg, D. The polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Bulut-Karslioglu, A. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 55, 277–290 (2014).
Nicetto, D. et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363, 294–297 (2019).
Wang, C. et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018).
Pace, L. et al. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359, 177–186 (2018).
Soufi, A., Donahue, G. & Zaret, K. S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012).
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Schultz, D. C., Ayyanathan, K., Negorev, D., Maul, G. G. & Rauscher, F. J. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002).
Johnson, W. L. et al. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6, e25299 (2017).
Zhang, K., Mosch, K., Fischle, W. & Grewal, S. I. S. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388 (2008).
Allshire, R. C. & Madhani, H. D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018).
Sugiyama, T. et al. Enhancer of rudimentary cooperates with conserved RNA-Processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 61, 747–759 (2016).
Xie, G. et al. A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing. Nat. Commun. 10, 251 (2019).
Hazra, D., Andrić, V., Palancade, B., Rougemaille, M. & Graille, M. Formation of S. pombe Erh1 homodimer mediates gametogenic gene silencing and meiosis progression. Sci. Rep. 10, 1034 (2020).
Djupedal, I. et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19, 2301–2306 (2005).
Kato, H. et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309, 467–469 (2005).
Onodera, Y. et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622 (2005).
Pinzón, N. et al. Functional lability of RNA-dependent RNA polymerases in animals. PLoS Genet. 15, e1007915 (2019).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Huang, P. et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384 (2014).
Liu, M.-L. et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4, 2183 (2013).
Cacchiarelli, D. et al. Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell 162, 412–424 (2015).
Hermann, B. P. et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650–1667 (2018).
Zhang, Y. et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol. Cell 72, 1021–1034 (2018).
Cheloufi, S. et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015).
Jeong, H.-H., Yalamanchili, H. K., Guo, C., Shulman, J. M. & Liu, Z. An ultra-fast and scalable quantification pipeline for transposable elements from next generation sequencing data. Pac. Symp. Biocomput. 23, 168–179 (2018).
Lewis, Z. A. et al. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 6, e1001196 (2010).
Kouprina, N. et al. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PLoS ONE 2, e359 (2007).
Bao, X. et al. Capturing the interactome of newly transcribed RNA. Nat. Methods 15, 213–220 (2018).
Caudron-Herger, M. et al. R-DeeP: proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol. Cell 75, 184–199 (2019).
Wang, C. et al. A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res. 44, 969–982 (2016).
Scarola, M. et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 6, 7631 (2015).
Kalyana-Sundaram, S. et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149, 1622–1634 (2012).
Criscione, S. W., Zhang, Y., Thompson, W., Sedivy, J. M. & Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 15, 583 (2014).
Zhu, Z. et al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 20, 794–801 (2010).
Peters, A. H. F. M. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003).
Patil, D. P. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Tchasovnikarova, I. A. et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485 (2015).
Yamazaki, T. et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053 (2018).
Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78, 554–565 (2020).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Liu, Y. et al. PWP1 mediates nutrient-dependent growth control through nucleolar regulation of ribosomal gene expression. Dev. Cell 43, 240–252 (2017).
Collier, S. P., Collins, P. L., Williams, C. L., Boothby, M. R. & Aune, T. M. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 189, 2084–2088 (2012).
Agarwal, V., Bell, G. W., Nam, J.-W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015).
Zhang, T. et al. FUS regulates activity of microRNA-mediated gene silencing. Mol. Cell 69, 787–801 (2018).
Villarroya-Beltri, C. et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980 (2013).
Yamazaki, T. & Hirose, T. The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. (Elite Ed.) 7, 1–41 (2015).
Reynolds, N. et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of polycomb repressive complex 2 to direct gene repression. EMBO J. 31, 593–605 (2012).
Chalamcharla, V. R., Folco, H. D., Dhakshnamoorthy, J. & Grewal, S. I. S. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl Acad. Sci. USA 112, 15548–15555 (2015).
Mattout, A. et al. LSM2-8 and XRN-2 contribute to the silencing of H3K27me3-marked genes through targeted RNA decay. Nat. Cell Biol. 22, 579–590 (2020).
Shima, H. et al. S-Adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363 (2017).
van Pelt, J. F., Decorte, R., Yap, P. S. H. & Fevery, J. Identification of HepG2 variant cell lines by short tandem repeat (STR) analysis. Mol. Cell Biochem 243, 49–54 (2003).
Jaccard, N. et al. Automated method for the rapid and precise estimation of adherent cell culture characteristics from phase contrast microscopy images. Biotechnol. Bioeng. 111, 504–517 (2014).
Huang, J. et al. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience 6, 1–9 (2017).
Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, 1–6 (2018).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Acknowledgements
R.L.M. was supported by an NIH K01 postdoctoral fellowship DK117970-01. K.E.K. was supported by an NIH T32 predoctoral training program in Developmental Biology HD083185 and the Blavatnik Family Fellowship in Biomedical Research. A.H. was supported by the Conquer Cancer, the ASCO Foundation Young Investigator Award and a National Institutes of Health training grant (T32 DK007066). S.H.K. and B.L. were supported by NIH R35 GM133425. The research was supported by NIH P01 GM099134 and two grants from the Pennsylvania Department of Health to K.S.Z.
Author information
Authors and Affiliations
Contributions
R.L.M., K.E.K. and K.S.Z. designed the study and wrote the manuscript. R.L.M., K.E.K., S.H.K., Y.Z., L.X., A.H., J.S.B. and O.A. conducted the experiments. Y.Z., L.X. and Y.H. performed the RNA-seq. R.L.M., K.E.K., Y.Z., L.X. and G.D. performed the bioinformatics analysis. S.H.K. and B.L. performed confocal imaging, quantification and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of siRNA efficiency and confirmation of RNA-seq results by qPCR.
a, qPCR quantification of knockdown for all siRNAs used in this study. siRNAs used for treating cells for RNA-seq analysis indicated in red. Above bar graphs show the number of srHC genes significantly upregulated (DESeq2, Benjamini multiple test corrected Wald test p-value ≤0.05 and log2(foldchange)>0) by each knockdown in reprogramming and non-reprogramming conditions. (n=2 biological replicates per siRNA, two siRNAs per target) b, Protein depletion efficiency for select srHC proteins in study, asterisk color corresponds to knockdown in (a). Arrows indicate location of indicated molecular weight markers. Experiment repeated independently 2 times with similar results. Unprocessed blots are provided as source data. c, Quantification of cell confluency using PHANTAST55 from phase contrast images (n=4 images for each condition; two independently targeting siRNAs per target, two replicates per siRNA). Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values.
Extended Data Fig. 2 Extended analysis of top noTF srHC proteins.
a, Sucrose gradient fractionation of sonicated DNA, DNA concentration of each fraction indicated (20ul loaded per lane), followed by western blot probing for ERH with RBMX and H3 as controls. Experiment repeated independently 2 times with similar results. b, qRT-PCR in siControl (n=3) and siERH treated (n=3 for each of two different siRNAs) human fibroblasts of pre-RNAs of genes upregulated at mRNA level by ERH depletion (two tailed Student’s t-test). Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. c, Spermatogenic TF motif enrichment in srHC spermatogenesis genes upregulated and not upregulated by each of the n=97 siRNA targets (two tailed Student’s t-test). Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. d, Western blots for transcription factors involved in spermatogenesis to assess levels in fibroblast whole cell lysate. e, Western blots for transcription factors with motifs enriched in promoters of srHC genes activated by ERH knockdown. f, Expanded Salmon TE heatmaps for knockdowns showing highest repeat activation (arrows indicate subtypes with the highest percent of upregulation by indicated knockdown). P values from b,c are denoted in the panels. Statistical information and unprocessed blots are provided as source data.
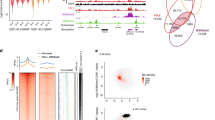
Extended Data Fig. 3 ERH depletion causes a global decrease in H3K9me3 but does not decrease H3K9 HMT expression or H3K9me2 levels.
a, Quantification of H3K9me3 immunofluorescence in siControl and siERH treated human fibroblasts (Student’s two tailed t-test). Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. N numbers are denoted in the panel and represent the number of cells imaged per treatment. b, Volcano plots showing expression change and significance of indicated H3K9 histone methyltransferase for 97 siRNA knockdowns by RNA-seq (n=2). siRNA knockdowns causing significant (DEseq2, Benjamini multiple test corrected Wald test p-value ≤0.05 and log2(foldchange)>0) upregulation (red) or downregulation (blue) are listed within graph. c, H3K9me3 immunofluorescence (left) and quantification (right) in siControl (n=352 cells) and siERH (n=365 cells) treated HepG2 cells (two tailed Student’s t-test). Experiment repeated independently 2 times with similar results. Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. P values denoted in panel. d, Western blots for H3K9me3 histone methyltransferases in siControl and siERH treated human fibroblasts. Experiment repeated independently 2 times with similar results. e, Western blots for ERH, SUV39H1 and H3K9me3 in the sonicated chromatin fraction from siControl and siERH treated human fibroblasts. Arrows indicate location of indicated molecular weight markers. Experiment repeated independently 2 times with similar results. f, H3K9me3 immunofluorescence comparison between siERH and HMTs siSUV39H1 and siSETDB1. Experiment repeated independently 2 times with similar results. g, H3K9me2 (green) and DAPI (blue) immunofluorescence in siControl and siERH treated human fibroblasts using an alternative antibody. Experiment repeated independently 2 times with similar results. h, H3K9me3 immunofluorescence in siControl and siSKIV2L2 treated human fibroblasts. Scale bars indicate 50µm for all images. Experiment repeated independently 2 times with similar results. Unprocessed blots are provided as source data.
Extended Data Fig. 4 ERH regulates gametogenic genes and a subset of repeat elements.
a, RNA-seq tracks showing SPANX cluster expression in siERH and several additional srHC protein knockdowns; same scale used for all mRNA-seq tracks. b, Protein sequence alignment of the ERH interacting domain of Mmi1 (95–122) and corresponding regions, determined by full length protein alignment, of the closest human orthologs. Purple arrow indicates tryptophan residue previously observed in a separate study2 to be important for ERH interaction. c, R-Deep database showing control and RNase treated fractionation and mass spectrometry detection of ERH. Other proteins observed to fractionate with ERH in fraction 22 which also exhibit a RNase induced shift listed in red box. d, Correlation of RepEnrich analysis of H3K9me3 and expression changes of repeat element classes in siERH relative to siControl. Alu and ERVK elements showed the greatest negative correlation between H3K9me3 and expression and are plotted separately. R correlation coefficient and p-value calculated using corrcoef in MATLAB; p-value calculated as the corresponding two-sided p-value for the t-distribution with n-2 degrees of freedom. N numbers stated in panel and represent the number of distinct repeat elements of indicated class. e, Violin plots of H3K9me3 and expression fold change (log2(siERH/siControl)) as determined by RepEnrich for the repeat element classes exhibiting significant H3K9me3 and expression changes (two tailed Student’s t-test). N numbers stated in panel and represent the number of distinct repeat elements of indicated class. f, H3K27me3 changes at satellite repeats in siERH relative to siControl. g, Motif occurrence for FOXA3, HNF1α and HNF4α in ERV (n=1641 element sequences), LINE (n=471 element sequences) and SINE (n=147 element sequences) elements (two tailed Student’s t-test). Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. h, Quantification of flow cytometry assay of siRNA+hiHep cells stained for the hepatic marker ASGPR1 (two tailed Student’s t-test to siControl). N numbers stated in panel and representthe number of cells quantified to calculate percent positive per condition. P values in d,e,g,h are denoted in the panels.
Extended Data Fig. 5 Chromosomal positions of srHC gene activation for noTF and hiHep.
Chromosomal positions for hiHep activation of srHC genes by indicated knockdowns. Expanded example regions 1–3 are marked by grey background.
Extended Data Fig. 6 Histone profiles of genes targeted by hub proteins.
a,b,c, H3K9me3 (a), H3K27me3 (b) and srHC (c) meta profiles of genes targeted by hub proteins 2.5kb upstream of TSS to 2.5kb downstream of TTS.
Extended Data Fig. 7 Extended cluster specific analysis.
a, Gene Ontology analysis for statistical overrepresentation of biological processes for srHC genes uniquely repressed by each cluster; non-redundant GO categories shown (p-value calculated by PANTHER Statistical overrepresentation test, denoted in panel). N numbers represent number of genes in GO category repressed by srHC protein member of indicated cluster and are stated in panel. b, Profiles of A/B compartment enrichment43, H4K20me1 and DNA methylation by bisulfide sequencing across genes uniquely regulate by each cluster. c, TargetScan database analysis for enrichment of miRNA target sequences in srHC genes uniquely repressed by each cluster; -log2(p-value) shown for the top 55 enriched miRNA target sequences per cluster (p-value calculated using the statistical model developed in Agarwal et al., 2017). d, Heatmap of H3K9me3, H3K27me3 and srHC-seq profiles of srHC genes uniquely regulated by each srHC protein cluster and sorted by mean H3K9me3 level within each cluster set. N numbers represent number of srHC gene profiles depicted and are stated in the panel.
Extended Data Fig. 8 srHC protein ChIP-seq.
a, ChIP-seq profiles of HMGA1 and MYBBP1A at srHC genes repressed by both factors or repressed by the other factor. b, Browser track showing ERH at a H3K9me3 marked euchromatin domain. c, srHC and euchromatin H3K9m3 gene expression changes in siERH+hiHep. N numbers represent the number of srHC or euchromatin srHC genes and are stated in the panel. The number of genes in each set significantly up (red) or down (blue) are indicated. d, DNA fragment size profiles by BioAnalyzer of INPUT and eluted DNA after ChIP of the indicated srHC protein. e, Table showing the percent of DNA fragments with length>=1kb as determined by paired-end sequencing for reads mapping with over 50% overlap with euchromatin or srHC domains (two tailed Student’s t-test, n=3 biological replicates sequenced as INPUT).
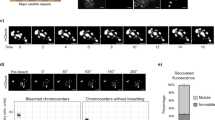
Extended Data Fig. 9 Quantification of confocal images and extended analysis.
a, Methodology for defining nuclear border and quantifying fluorescence intensity. b, H3K9me3 and H3K27me3 immunofluorescence in siControl and siXRN2 treated human fibroblasts. Images representative of 2 experiments. c, H3K9me3 immunofluorescence in siControl and siYTHDC1 treated human fibroblasts. Images representative of 4 experiments. d,e Representative images (d) and quantification (e) of H3K27me3 for cells depleted by siRNA for hub proteins. Images representative of 2 experiments. Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. N numbers for e represent the number of cells imaged and are stated in the panel. f, Plot showing correlation of H3K9me3 immunofluorescence intensity relative to siControl vs the number of hepatic srHC genes induced during hiHep reprogramming for the target srHC proteins in Fig. 5b. R correlation coefficient and p-value calculated using corrcoef in MATLAB; p-value calculated as the corresponding two-sided p-value for the t-distribution with n-2 degrees of freedom. N numbers stated in panel and represent the number of siRNA targets. All scale bars indicate 50 µm.
Extended Data Fig. 10 Expanded analysis of ChIP-seq and srHC-seq in siERH.
a, DNA size profiles of sonicated fractions from siControl and siERH. b, Combinatorial H3K9me3 and H3K27me3 levels for srHC genes in siControl treated human fibroblasts. c, Heatmap displaying enrichment of H3K9me3, H3K27me3, dual-marked, and unmarked srHC gene subtypes in siControl and siERH from 2kp upstream of TSS to 2kp downstream of TTS. d, Activation of srHC alternative lineage genes in siERH relative to siControl during hiHep reprogramming not activated by siERH without hiHep factors. e, Percent occurrence of top 5 motifs enriched in promoters of non-hepatic genes upregulated in siERH+hiHep conditions and corresponding expression of the putative targeting factors in 4 siControl+hiHep replicates. f, H3K9me3 changes at classes of key hepatic genes, cytochrome p450 (n=50), UGT (n=21), SLC transporter (n=196) and ABC transporter (n=27), in siERH compared to siControl. Boxplot center, bounds and whiskers represent the median, 25–75% range and minimum to maximum values. g,h, Location of srHC genes gaining H3K9me3 (g) and H3K27me3 (h) on t-SNE embedding. i, hiHep motifs from Jaspar database and identified motifs in promoter regions (tss +/− 200) of srHC genes with motif scores >=10. j, Table showing total gene numbers and activation rates in siERH for sets of srHC genes defined by presence of strong hiHep motifs and specific changes in H3K9me3 and H3K27me3 levels.
Supplementary information
Supplementary Information
Supplementary Fig. 1: chromatin features of representative genes targeted by each srHC protein cluster. a–d, Browser tracks for example srHC genes repressed by cluster 1 (a), cluster 2 (b), cluster 3 (c) or cluster 4 (d) and no other clusters.
Supplementary Tables
Supplementary Table 1: table of the siRNAs used in study. Supplementary Table 2: table of the RT–qPCR primers used in study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data for Fig. 3a,b.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots of Extended Data Fig. 1b.
Source Data Extended Data Fig. 2
Unprocessed western blots of Extended Fig. 2a and western blots of Extended Data Fig. 2d,e.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots of Extended Data Fig. 3c,d.
Rights and permissions
About this article
Cite this article
McCarthy, R.L., Kaeding, K.E., Keller, S.H. et al. Diverse heterochromatin-associated proteins repress distinct classes of genes and repetitive elements. Nat Cell Biol 23, 905–914 (2021). https://doi.org/10.1038/s41556-021-00725-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00725-7
This article is cited by
-
RepEnTools: an automated repeat enrichment analysis package for ChIP-seq data reveals hUHRF1 Tandem-Tudor domain enrichment in young repeats
Mobile DNA (2024)
-
Heterochromatin definition and function
Nature Reviews Molecular Cell Biology (2023)
-
Germinal center output is sustained by HELLS-dependent DNA-methylation-maintenance in B cells
Nature Communications (2023)
-
Collateral activity of the CRISPR/RfxCas13d system in human cells
Communications Biology (2023)
-
HMGA2 directly mediates chromatin condensation in association with neuronal fate regulation
Nature Communications (2023)