Abstract
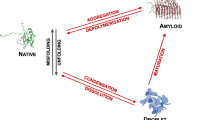
Proteins undergoing liquid–liquid phase separation are being discovered at an increasing rate. Since at the high concentrations present in the cell most proteins would be expected to form a liquid condensed state, this state should be considered to be a fundamental state of proteins along with the native state and the amyloid state. Here we discuss the generic nature of the liquid-like and solid-like condensed states, and describe a wide variety of biological functions conferred by these condensed states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Knowles, T. P., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014).
Vecchi, G. et al. Proteome-wide observation of the phenomenon of life on the edge of solubility. Proc. Natl Acad. Sci. USA 117, 1015–1020 (2020).
Hardenberg, M., Horvath, A., Ambrus, V., Fuxreiter, M. & Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl Acad. Sci. USA 117, 33254–33262 (2020).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Brangwynne, C. P., Tompa, P. & Pappu, R. V. Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 (2015).
Alberti, S. The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 130, 2789–2796 (2017).
Tartaglia, G. G., Pechmann, S., Dobson, C. M. & Vendruscolo, M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 32, 204–206 (2007).
Sormanni, P. et al. Simultaneous quantification of protein order and disorder. Nat. Chem. Biol. 13, 339–342 (2017).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Sawaya, M. R. et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Reichheld, S. E., Muiznieks, L. D., Keeley, F. W. & Sharpe, S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl Acad. Sci. USA 114, E4408–E4415 (2017).
Blake, C. C., Mair, G. A., North, A. C., Phillips, D. C. & Sarma, V. R. On the conformation of the hen egg-white lysozyme molecule. Proc. R. Soc. Lond. B 167, 365–377 (1967).
Wilson, K. P. et al. Structure and mechanism of interleukin-1β converting enzyme. Nature 370, 270–275 (1994).
Fuxreiter, M. & Warshel, A. Origin of the catalytic power of acetylcholineesterase. Computer simulation studies. J. Am. Chem. Soc. 120, 183–194 (1998).
Aggarwal, A. K., Rodgers, D. W., Drottar, M., Ptashne, M. & Harrison, S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242, 899–907 (1988).
Slupphaug, G. et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 (1996).
Hosfield, D. J., Guan, Y., Haas, B. J., Cunningham, R. P. & Tainer, J. A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98, 397–408 (1999).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Ciechanover, A. & Schwartz, A. L. How are substrates recognized by the ubiquitin-mediated proteolytic system? Trends Biochem. Sci. 14, 483–488 (1989).
Song, L. et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996).
Takizawa, M. et al. Mechanistic basis for the recognition of laminin-511 by α6β1 integrin. Sci. Adv. 3, e1701497 (2017).
Steitz, T. A. Visualizing polynucleotide polymerase machines at work. EMBO J. 25, 3458–3468 (2006).
Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002).
Manglik, A. et al. Structural Insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161, 1101–1111 (2015).
Cheng, X., Kumar, S., Posfai, J., Pflugrath, J. W. & Roberts, R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell 74, 299–307 (1993).
Jiang, Y., Rossi, P. & Kalodimos, C. G. Structural basis for client recognition and activity of Hsp40 chaperones. Science 365, 1313–1319 (2019).
Tompa, P. & Fuxreiter, M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33, 2–8 (2008).
Selenko, P. et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11, 965–976 (2003).
Chaudhry, C. et al. Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J. 22, 4877–4887 (2003).
Saleh, T., Rossi, P. & Kalodimos, C. G. Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol. 24, 893–901 (2017).
West, A., Brummel, B. E., Braun, A. R., Rhoades, E. & Sachs, J. N. Membrane remodeling and mechanics: Experiments and simulations of α-synuclein. Biochim. Biophys. Acta 1858, 1594–1609 (2016).
Yuen, F. et al. Preferential adsorption to air-water interfaces: a novel cryoprotective mechanism for LEA proteins. Biochem. J. 476, 1121–1135 (2019).
Delaforge, E. et al. Deciphering the dynamic interaction profile of an intrinsically disordered protein by NMR exchange spectroscopy. J. Am. Chem. Soc. 140, 1148–1158 (2018).
Rowell, J. P., Simpson, K. L., Stott, K., Watson, M. & Thomas, J. O. HMGB1-facilitated p53 DNA binding occurs via HMG–Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 20, 2014–2024 (2012).
Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2020).
Freeman Rosenzweig, E. S. et al. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162 (2017).
Fowler, D. M. et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 4, e6 (2006).
Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016).
Schwarz-Romond, T., Metcalfe, C. & Bienz, M. Dynamic recruitment of axin by Dishevelled protein assemblies. J. Cell Sci. 120, 2402–2412 (2007).
Woodruff, J. B. et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017).
Banjade, S. & Rosen, M. K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. 3, e04123 (2014).
Case, L. B., Zhang, X., Ditlev, J. A. & Rosen, M. K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 (2019).
Du, M. & Chen, Z. J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018).
Li, J. et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 (2012).
Kilic, S. et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379 (2019).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018).
Larson, A. G. et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Zaffagnini, G. et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308 (2018).
Fujioka, Y. et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305 (2020).
Claessen, D. et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726 (2003).
Kwan, A. H. et al. Structural basis for rodlet assembly in fungal hydrophobins. Proc. Natl Acad. Sci. USA 103, 3621–3626 (2006).
Quiroz, F. G. et al. Liquid–liquid phase separation drives skin barrier formation. Science 367, eaax9554 (2020).
Iconomidou, V. A., Vriend, G. & Hamodrakas, S. J. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 479, 141–145 (2000).
Boke, E. et al. Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 (2016).
Maji, S. K. et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 (2009).
Chapman, M. R. et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002).
Dueholm, M. S. et al. Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 (2010).
Miskei, M. et al. Fuzziness enables context dependence of protein interactions. FEBS Lett. 591, 2682–2695 (2017).
Wippich, F. et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013).
Milles, S. et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 163, 734–745 (2015).
Frottin, F. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Krishnan, R. & Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005).
Baxa, U., Ross, P. D., Wickner, R. B. & Steven, A. C. The N-terminal prion domain of Ure2p converts from an unfolded to a thermally resistant conformation upon filament formation. J. Mol. Biol. 339, 259–264 (2004).
Ghosh, S. et al. p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 24, 1784–1798 (2017).
Lin, S. C., Lo, Y. C. & Wu, H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 (2010).
Cai, X. et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222 (2014).
Chakrabortee, S. et al. Intrinsically disordered proteins drive emergence and inheritance of biological traits. Cell 167, 369–381.e12 (2016).
Shi, Y. et al. An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat. Commun. 6, 7811 (2015).
Hou, F. et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461 (2011).
Itakura, A. K., Chakravarty, A. K., Jakobson, C. M. & Jarosz, D. F. Widespread prion-based control of growth and differentiation strategies in saccharomyces cerevisiae. Mol. Cell 77, 266–278 (2020).
Zhao, Y., Chen, S., Swensen, A. C., Qian, W. J. & Gouaux, E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362 (2019).
Milovanovic, D., Wu, Y., Bian, X. & De Camilli, P. A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 (2018).
Zeng, M. et al. Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell 174, 1172–1187.e16 (2018).
Heinrich, S. U. & Lindquist, S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB). Proc. Natl Acad. Sci. USA 108, 2999–3004 (2011).
Kirschner, M. & Gerhart, J. Evolvability. Proc. Natl Acad. Sci. USA 95, 8420–8427 (1998).
Gianni, S. et al. Fuzziness and frustration in the energy landscape of protein folding, function, and assembly. Acc. Chem. Res. 54, 1251–1259 (2021).
Miskei, M., Horvath, A., Vendruscolo, M. & Fuxreiter, M. Sequence-based prediction of fuzzy protein interactions. J. Mol. Biol. 432, 2289–2303 (2020).
Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015).
Burke, K. A., Janke, A. M., Rhine, C. L. & Fawzi, N. L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 (2015).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation–pi interactions. Cell 173, 720–734.e15 (2018).
Ray, S. et al. α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 12, 705–716 (2020).
Wegmann, S. et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018).
Niaki, A. G. et al. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol. Cell 77, 82–94 (2020).
Kim, T. H. et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 (2019).
Gibson, B. A. et al. Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Patel, A. et al. ATP as a biological hydrotrope. Science 356, 753–756 (2017).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019).
Jang, S. et al. Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron 90, 278–291 (2016).
Jang, S. et al. Phosphofructokinase relocalizes into subcellular compartments with liquid-like properties in vivo. Biophys. J. 120, 1170–1186 (2021).
Wu, H. & Fuxreiter, M. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 165, 1055–1066 (2016).
Alberti, S. & Dormann, D. Liquid–liquid phase separation in disease. Annu Rev. Genet. 53, 171–194 (2019).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Hardenberg, M. C. et al. Observation of an α-synuclein liquid droplet state and its maturation into Lewy body-like assemblies. J. Mol. Cell Biol. https://doi.org/10.1093/jmcb/mjaa075 (2021).
Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021).
Marrone, L., Quamar, S., Mannini, B., St George-Hyslop, P. & Vendruscolo, M. P525L promotes the aggregation of FUS by altering its biochemical and biophysical properties. Science Matters https://sciencematters.io/articles/202004000008/info (2020).
Markmiller, S. et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604 (2018).
Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345 (2020).
Li, W. et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 22, 960–972 (2020).
Vendruscolo, M. & Fuxreiter, M. Sequence determinants of the aggregation of proteins within condensates generated by liquid-liquid phase separation. Preprint at BioRxiv https://doi.org/10.1101/2020.12.07.414409 (2020).
Guo, L. et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692 (2018).
Heller, G. T. et al. Small-molecule sequestration of amyloid-beta as a drug discovery strategy for Alzheimer’s disease. Sci. Adv. 6, eabb5924 (2020).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Song, H., Inaka, K., Maenaka, K. & Matsushima, M. Structural changes of active site cleft and different saccharide binding modes in human lysozyme co-crystallized with hexa-N-acetyl-chitohexaose at pH 4.0. J. Mol. Biol. 244, 522–540 (1994).
Huang, W. Y. C. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019).
Nogales, E. An electron microscopy journey in the study of microtubule structure and dynamics. Protein Sci. 24, 1912–1919 (2015).
Huang, D. T. et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature 445, 394–398 (2007).
Park, J. E. et al. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat. Commun. 10, 4959 (2019).
Wasmer, C. et al. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 (2008).
Ciuffa, R. et al. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 11, 748–758 (2015).
Song, H. & Ji, X. The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun. 10, 3085 (2019).
Gross, J. D. et al. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115, 739–750 (2003).
Cao, Q., Boyer, D. R., Sawaya, M. R., Ge, P. & Eisenberg, D. S. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol. 26, 619–627 (2019).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fuxreiter, M., Vendruscolo, M. Generic nature of the condensed states of proteins. Nat Cell Biol 23, 587–594 (2021). https://doi.org/10.1038/s41556-021-00697-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00697-8
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
MolPhase, an advanced prediction algorithm for protein phase separation
The EMBO Journal (2024)
-
Amyloid formation as a protein phase transition
Nature Reviews Physics (2023)
-
Alternatively spliced exon regulates context-dependent MEF2D higher-order assembly during myogenesis
Nature Communications (2023)
-
Formation of nuclear CPSF6/CPSF5 biomolecular condensates upon HIV-1 entry into the nucleus is important for productive infection
Scientific Reports (2023)