Abstract
Coordination of stem cell function by local and niche-derived signals is essential to preserve adult tissue homeostasis and organismal health. The vasculature is a prominent component of multiple stem cell niches. However, its role in adult intestinal homeostasis remains largely understudied. Here we uncover a previously unrecognised crosstalk between adult intestinal stem cells in Drosophila and the vasculature-like tracheal system, which is essential for intestinal regeneration. Following damage to the intestinal epithelium, gut-derived reactive oxygen species activate tracheal HIF-1α and bidirectional FGF/FGFR signalling, leading to reversible remodelling of gut-associated terminal tracheal cells and intestinal stem cell proliferation following damage. Unexpectedly, reactive oxygen species-induced adult tracheal plasticity involves downregulation of the tracheal specification factor trachealess (trh) and upregulation of IGF2 messenger RNA-binding protein (IGF2BP2/Imp). Our results reveal an intestine–vasculature inter-organ communication programme that is essential to adapt the stem cell response to the proliferative demands of the intestinal epithelium.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
TaDa sequencing data including all raw sequence files and processed files have been deposited in the Gene Expression Omnibus under the accession number GSE163570. Public databases used in this study include: FlyBase (https://flybase.org) and ftp://ftp.flybase.net/genomes/Drosophila_melanogaster/. Source data are provided with this paper. Any additional data supporting this study will be made available by the corresponding author on reasonable request.
Code availability
The custom Perl script used for TaDa analysis47 is available at https://github.com/tonysouthall/Dam-RNA_POLII_analysis. The custom ImageJ macros used in this study are available at https://github.com/emltwc/TracheaProject. Any additional information on codes is available from the corresponding author on request.
References
Beumer, J. & Clevers, H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143, 3639–3649 (2016).
McCarthy, N., Kraiczy, J. & Shivdasani, R. A. Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol. 22, 1033–1041 (2020).
Hageman, J. H. et al. Intestinal regeneration: regulation by the microenvironment. Dev. Cell 54, 435–446 (2020).
Ghabrial, A., Luschnig, S., Metzstein, M. M. & Krasnow, M. A. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19, 623–647 (2003).
Hayashi, S. & Kondo, T. Development and function of the Drosophila tracheal system. Genetics 209, 367–380 (2018).
Eilken, H. M. & Adams, R. H. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr. Opin. Cell Biol. 22, 617–625 (2010).
Affolter, M. et al. Tube or not tube: remodeling epithelial tissues by branching morphogenesis. Dev. Cell 4, 11–18 (2003).
Cela, C. & Llimargas, M. Egfr is essential for maintaining epithelial integrity during tracheal remodelling in Drosophila. Development 133, 3115–3125 (2006).
Centanin, L., Gorr, T. A. & Wappner, P. Tracheal remodelling in response to hypoxia. J. Insect Physiol. 56, 447–454 (2010).
Linneweber, G. A. et al. Neuronal control of metabolism through nutrient-dependent modulation of tracheal branching. Cell 156, 69–83 (2014).
Casali, A. & Batlle, E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell 4, 124–127 (2009).
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006).
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006).
Miguel-Aliaga, I., Jasper, H. & Lemaitre, B. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics 210, 357–396 (2018).
Li, W. et al. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J. Neurosci. 33, 5821–5833 (2013).
Sutherland, D., Samakovlis, C. & Krasnow, M. A. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87, 1091–1101 (1996).
Guillemin, K. et al. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development 122, 1353–1362 (1996).
Gervais, L. & Casanova, J. The Drosophila homologue of SRF acts as a boosting mechanism to sustain FGF-induced terminal branching in the tracheal system. Development 138, 1269–1274 (2011).
Centanin, L. et al. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev. Cell 14, 547–558 (2008).
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009).
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S. & Lemaitre, B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211 (2009).
Buchon, N., Broderick, N. A., Chakrabarti, S. & Lemaitre, B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344 (2009).
Apidianakis, Y., Pitsouli, C., Perrimon, N. & Rahme, L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl Acad. Sci. USA 106, 20883–20888 (2009).
Morel, F., Renoux, M., Lachaume, P. & Alziari, S. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat. Res. 637, 111–117 (2008).
Amcheslavsky, A., Jiang, J. & Ip, Y. T. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49–61 (2009).
Tian, A., Wang, B. & Jiang, J. Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc. Natl Acad. Sci. USA 114, E2699–E2708 (2017).
Ren, F. et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl Acad. Sci. USA 107, 21064–21069 (2010).
Kim, C. K., Yang, V. W. & Bialkowska, A. B. The role of intestinal stem cells in epithelial regeneration following radiation-induced gut injury. Curr. Stem Cell Rep. 3, 320–332 (2017).
Chatterjee, M. & Ip, Y. T. Pathogenic stimulation of intestinal stem cell response in Drosophila. J. Cell. Physiol. 220, 664–671 (2009).
Lavista-Llanos, S. et al. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell Biol. 22, 6842–6853 (2002).
Li, J., Hobman, T. C. & Simmonds, A. J. Gawky (GW) is the Drosophila melanogaster GW182 homologue. Adv. Exp. Med Biol. 768, 127–145 (2013).
Cordero, J. B., Stefanatos, R. K., Scopelliti, A., Vidal, M. & Sansom, O. J. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 31, 3901–3917 (2012).
Ha, E. M. et al. An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell 8, 125–132 (2005).
Hochmuth, C. E., Biteau, B., Bohmann, D. & Jasper, H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8, 188–199 (2011).
Rey, S. & Semenza, G. L. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc. Res. 86, 236–242 (2010).
Bacon, N. C. et al. Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1α. Biochem. Biophys. Res. Commun. 249, 811–816 (1998).
Chandel, N. S. et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1α during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 275, 25130–25138 (2000).
Ohshiro, T. & Saigo, K. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development 124, 3975–3986 (1997).
Horowitz, A. & Simons, M. Branching morphogenesis. Circ. Res. 103, 784–795 (2008).
Du, L. et al. Unique patterns of organization and migration of FGF-expressing cells during Drosophila morphogenesis. Dev. Biol. 427, 35–48 (2017).
Johansson, J. et al. RAL GTPases drive intestinal stem cell function and regeneration through internalization of WNT signalosomes. Cell Stem Cell 24, 592–607 e597 (2019).
Muha, V. & Muller, H. A. Functions and mechanisms of fibroblast growth factor (FGF) signalling in Drosophila melanogaster. Int. J. Mol. Sci. 14, 5920–5937 (2013).
Biteau, B. & Jasper, H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045–1055 (2011).
Buchon, N., Broderick, N. A., Kuraishi, T. & Lemaitre, B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 8, 152 (2010).
Xu, N. et al. EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev. Biol. 354, 31–43 (2011).
Marshall, O. J., Southall, T. D., Cheetham, S. W. & Brand, A. H. Cell-type-specific profiling of protein–DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat. Protoc. 11, 1586–1598 (2016).
Southall, T. D. et al. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 26, 101–112 (2013).
Sandoval, J. et al. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 32, e88 (2004).
Medioni, C., Ramialison, M., Ephrussi, A. & Besse, F. Imp promotes axonal remodeling by regulating profilin mRNA during brain development. Curr. Biol. 24, 793–800 (2014).
Samuels, T. J., Jarvelin, A. I., Ish-Horowicz, D. & Davis, I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. eLife 9, e51529 (2020).
Chung, S., Chavez, C. & Andrew, D. J. Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev. Biol. 360, 160–172 (2011).
Isaac, D. D. & Andrew, D. J. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 10, 103–117 (1996).
Wilk, R., Weizman, I. & Shilo, B. Z. trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 10, 93–102 (1996).
Tamamouna et al. Remodelling of oxygen-transporting tracheoles drives intestinal regeneration and tumorigenesis in Drosophila. Nat. Cell Biol. (2021).
Holloway, E. M. et al. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Dev. Cell 54, 516–528 (2020).
Pasquier, J. et al. Angiocrine endothelium: from physiology to cancer. J. Transl. Med. 18, 52 (2020).
Dai, N. et al. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab. 21, 609–621 (2015).
Zhou, S., Degan, S., Potts, E. N., Foster, W. M. & Sunday, M. E. NPAS3 is a trachealess homolog critical for lung development and homeostasis. Proc. Natl Acad. Sci. USA 106, 11691–11696 (2009).
Nusse, Y. M. et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113 (2018).
Poplawski, G. H. D. et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature 581, 77–82 (2020).
Yui, S. et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49 (2018).
Marshall, O. J. & Brand, A. H. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics 31, 3371–3373 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Acknowledgements
J.P. and J.B.C. are funded by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (grant no. 104103/Z/14/Z; J.B.C.) and a Wellcome Trust Institutional Strategic Support Fund (ISSF)−Excellence and Innovation Catalyst Award to J.B.C. J.P. was partly funded by a BBSRC−Flexible Talent Mobility Account (FTMA) Award (grant no. BB/R506576/1). Y.Y. and A.B.M. are funded by CRUK core funding to the CRUK Beatson Institute (grant no. A17196). T.D.S. and G.N.A. were funded by a Wellcome Trust Investigator grant (grant no. 104567; T.D.S.) and a BBSRC grant (grant no. BB/P017924/1; T.D.S. and G.N.A.). We thank M. Llimargas, M. Affolter, I. Miguel-Aliaga, A. Brand, C. Polesello, A. Scopelliti, P. Wappner, I. Davis, F. Besse, H. Stoker, G. Morata and F. H. Dion for generously sharing reagents and fly lines. We thank the Kyoto, Vienna and Bloomington Drosophila Stock Centres and the Drosophila Studies Hybridoma Bank for fly stocks and antibodies. We thank C. Nixon (Beatson CRUK histology service) for immunohistochemistry of mouse intestinal samples, E. McKenzie for help and training on the use of the hypoxia chamber, M. Mullin for assistance with transmission electron microscopy, and D. Strachan, J. Halpin and R. Insall (Beatson CRUK) for their support with image quantification. We thank D. McGuinness and J. Galbraith (Glasgow Polyomics) for sequencing samples for TaDa and R. Stefanatos for advice with RT–qPCR. We thank M. Naszai for help with the creation of custom ImageJ macros and L. Carroll for providing mouse intestinal samples. We thank J.-P. Parvy and multiple members of the Cordero laboratory for scientific discussion and advice on the project.
Author information
Authors and Affiliations
Contributions
J.P. designed and carried out most experiments and analysed and interpreted the data. Y.Y. provided technical support throughout the study and performed RT–qPCR. J.P., G.N.A., T.D.S. and J.B.C. analysed the TaDa data. A.B.M. performed the experiments in Extended Data Fig. 5k–p. J.B.C. conceived the project, designed experiments, analysed the data and supervised the study. J.P. and J.B.C. wrote the paper with contributions from the rest of the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Rongwen Xi, Yiorgos Apidianakis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization and quantification of adult tracheal remodelling following intestinal damage.
a, (top), Adult posterior midgut and associated TTCs (green). (bottom), illustration of the different steps followed for the quantification of gut tracheal coverage as explained in Methods. Scale bar: 100 µm. Box in top panel highlights the area shown in the bottom panels b, Confocal images of TTCs (green) from posterior midguts of control animals (Suc) or animals fed with Bleomycin or DSS. Scale bar: 50 µm. c, Representative confocal images of Sucrose or Pe treated midguts (shaded in brown). Scale bar: 100 µm. d, e, Quantification of posterior midgut area (d) and ratio of tracheal coverage over adult posterior midgut area (e). Two-tailed, unpaired T-test; n = number of posterior midguts, indicated in panels. d, P = 0.3464. e, (****P < 0.0001). f, Confocal images of TTCs (green) from Sucrose or Pe treated midguts stained with anti-PH3 to label proliferating ISCs (red). Bottom panels represent orthogonal views of the midguts shown in top panels. Scale bar: 50 µm. g, Quantification of individual or combined TTC nuclei and PH3+ cells in control or Pe-infected midguts. Two-tailed, unpaired T-test (****P < 0.0001); n = number of posterior midguts, indicated in panel. h, Confocal images of adult TTCs assessed at the indicated time points during and after intestinal damage. White lines trace individual TTCs. Scale bar: 50 µm. i, Correlation graph between TTC coverage and ISC proliferation for each of the time points and conditions presented in (h). n = number of posterior midguts, indicated in panels. Pearson’s correlation coefficient (damage r = 0.9660, recovery r =recovery 0.9962, damage **P = 0.0075, recovery **P = 0.0038). Values represent mean ± s.e.m.
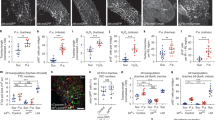
Extended Data Fig. 2 Hypoxia or impaired TTC remodelling does not affect ISC survival.
a, TTCs (green) and the Sima/HIF-1α activity reporter ldh-lacZ (grey) in adult posterior midguts from animals in normoxia (21% O2) or subjected to 24 hrs of hypoxia (3% O2). Dotted boxes show a magnified view of TTCs pointed by arrows. Scale bars: 50 µm (main figure); 20 µm (close up view). b, ldh-lacZ staining intensity within TTCs, relative to background, in midguts as in (a). Two-tailed, unpaired T-test (****P < 0.0001); n = number of TTCs from 9 posterior midguts per condition, indicated in panels. c, Tracheal coverage in adult posterior midguts as in (a). Two-tailed, unpaired T-test (***P = 0.0003); n = number of posterior midguts, indicated in panels. d, e, Midguts stained with anti-Dcp1 (magenta) to visualize cell death in control animals; upon adult-specific bax overexpression in ECs (d); animals subjected to hypoxia or to indicate the genetic TTC disruptions (e). d, e, Upper panels (apical tissue views); lower panels (longitudinal sections showing intestinal tube lumen). Scale bar: 50 µm. The high level of Dcp1 staining in the lumen of midguts subject to bax overexpression in ECs (d) corresponds to delaminating/dying cells. Dotted box in (e) shows a magnified view of an apoptotic EC pointed by arrow in main figure and identified by its large nuclei and Dcp1 staining. Scale bars: 50 µm (main figure); 20 µm (close up view). f, g, Quantification of Dcp1 staining in midguts as in (d, e). Two-tailed, unpaired T-test; n = number of posterior midguts, indicated in panels. f, (***P = 0.0009). g, (bax vs Control, ***P = 0.0003). h, TTCs (green) and ISCs detected with a Delta-lacZ reporter (magenta) and anti-Armadillo (Arm) staining (grey) in midguts as in (e). Dotted boxes in middle panels indicate the magnified areas in the lower panels. Scale bars: 50 µm (main figure); 20 µm (close up view). i, Number of Delta-lacZ positive cells in midguts as in (h). Two-tailed, unpaired T-test; n = number of midguts, indicates in panels. Values represent mean ± s.e.m.
Extended Data Fig. 3 Gut-associated TTCs are responsive to local signals from the damaged intestinal epithelium.
a, Examples of tracheal branching levels assigned to each of the scores used for the quantification of tracheal coverage from light-microscopy images. Scale bar: 50 µm. b, c (top panels), Quantification of tracheal branching from confocal (b) and bright-field images (c) of Sucrose or Pe treated midguts. Two-tailed, unpaired T-test (****P < 0.0001); n = number of posterior midguts, indicated in panels. b, c (bottom panels) Representative confocal (b) or bright-field images (c) from midguts as in top panels. Scale bars: 50 µm. d, Bright-field images from Sucrose or Pe treated midguts from wild-type (control) animals or upon catalase overexpression within ECs. Scale bar: 50 µm. e, f, Scoring of tracheal branching (e) and quantification of PH3+ ISCs (f) in posterior midguts as in (d). Two-way ANOVA followed by Sidak’s multiple comparisons test (****P < 0.0001); n = number of midguts quantified, indicated in panels. g, Bright-field images of posterior midgut from control animals or upon adult-specific overexpression of bax in ECs. Scale bar: 50 µm. h, i, Quantification of tracheal branching (h) and PH3+ ISCs (i) in posterior midguts as in (g). Two-tailed, unpaired T-test (****P < 0.0001); n = number of midguts, indicated in panels. Box plots represent maxima, minima and mean. Mean value is indicated on top of boxes. Otherwise, values represent mean ± s.e.m.
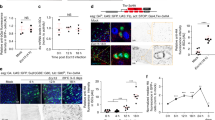
Extended Data Fig. 4 blistered/dSRF expression is upregulated in adult TTCs following intestinal damage or hypoxia and it regulates damage-induced TTC remodelling and ISC proliferation.
a, qRT-PCR for blistered (bs)/dSRF mRNA expression from adult midguts and associated tracheal tissue, in control (Sucrose) or Pe treated midguts. Two-tailed, unpaired T-test (*P = 0.0117); n = 5 biological replicates. b, Confocal images of bs-lacZ reporter expression (red) in Sucrose and Pe treated midguts stained with Chitin Binding Protein (CBP, green) to visualize all tracheal tissue. Dotted boxes show a magnified view of TTCs pointed by arrows. Scale bars: 50 µm (main figure); 20 µm (close up view). c, Quantification of bs-lacZ staining intensity within TTCs in posterior midguts as in (b). Two-tailed, unpaired T-test (****P < 0.0001); n = number of TTCs from 12-15 midguts per condition, indicated in panel. d, Confocal images of bs-lacZ reporter expression (red) in adult posterior midguts from animals housed in normoxia (21% O2) or subjected to 16 hrs of hypoxia (3% O2) and stained with CBP (green). Dotted boxes show a magnified view of TTCs pointed by arrows. Scale bars: 50 µm (main figure); 20 µm (close up view). e, Quantification of bs-lacZ staining intensity within TTCs in posterior midguts as in (d). Two-tailed, unpaired T-test (****P < 0.0001); n = number of TTCs from 12-15 midguts per condition, indicated in panel. f, Confocal images of Sucrose and Pe treated midguts from wild-type (control) animals or following RNAi-driven bs knockdown (bs−IR) within TTCs. Scale bar: 100 µm. g, h, Quantification of tracheal coverage (g) and PH3+ ISCs in posterior midguts as in (f). Two-way ANOVA followed by Sidak’s multiple comparisons test. g, (****P < 0.0001, *P < 0.05). h, (****P < 0.0001); n = number of posterior midguts quantified, indicated in panels. Values represent mean ± s.e.m.
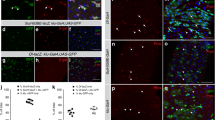
Extended Data Fig. 5 Multiple sources of Bnl individually contribute to regenerative ISC proliferation in the adult Drosophila midgut, independently of tracheal remodelling.
a, Confocal images of FGF/bnl reporter expression bnl > GFPnlacZ (green) in Sucrose and Pe treated midguts stained with CBP (magenta) to visualize all tracheal tissue and PH3 (magenta). Dotted boxes in left panels indicate the area magnified in the right panels. Scale bars: 100 µm (main figure); 40 µm (close up view). b, bnl > GFPnlacZ staining intensity relative to background within TTCs in midguts as in (a). Two-tailed, unpaired T-test (****P < 0.0001); n = number of TTCs from 12 midguts per condition, indicated in panel. c, d Tracheal coverage (c) and PH3+ ISCs (d) from wild-type (control) animals or upon bnl overexpression within TTCs. Two-tailed, unpaired T-test (****P < 0.0001); n = number of midguts, indicated in panels. e, Bright-field images from Sucrose or Pe treated midguts from wild-type (control) animals or following RNAi-driven bnl knockdown (bnl−IR) within ISCs/EBs. Scale bar: 50 µm. f, g, Tracheal branching (f) and PH3+ ISCs (g) in midguts as in (e). Two-way ANOVA followed by Sidak’s multiple comparisons test (****P < 0.0001); n = number of midguts, indicated in panels. h, Bright-field images from Sucrose or Pe treated midguts from wild-type (control) animals or upon bnl knockdown (bnl−IR) within ECs. Scale bar: 50 µm. i, j, Tracheal branching (i) and PH3+ ISCs (j) in midguts as in (h). Two-way ANOVA followed by Sidak’s multiple comparisons test (****P < 0.0001); n = number of midguts, indicated in panels. k, n, Bright-field images of midguts from wild-type (control) animals or upon bnl overexpression (bnl) within ISCs/EBs (k) or ECs (n). Scale bar: 50 µm. l, m, o, p, Tracheal coverage (l, o) and PH3+ ISCs (m, p) from midguts as in (k, n). Two-tailed, unpaired T-test; n = number of midguts, indicated in panels. l, (***P = 0.0002). m, (**P = 0.0066). o, (*P = 0.0213). p, (**P = 0.0084). Box plots represent maxima, minima and mean. Mean value is indicated on top of boxes. Otherwise, values represent mean ± s.e.m.
Extended Data Fig. 6 Individual sources of gut-derived Bnl are redundant for TTC remodelling but required for damage-induced ISC proliferation.
a, Bright-field images of Sucrose or Pe treated midguts from wild-type (control) animals or upon RNAi-driven btl knockdown (btl−IR) within ISCs/EBs. Scale bar: 50 µm. b, c, Quantification of tracheal branching (b) and PH3+ ISCs (c) in posterior midguts as in (a). Two-way ANOVA followed by Sidak’s multiple comparisons test (****P < 0.0001); n = number of posterior midguts, indicated in panels. d, Representative confocal images of activated MAPK (pERK) staining (red), Arm (grey) and ISCs/EBs (esg > GFP, green) in control (Sucrose) or regenerating (Pe) adult posterior midguts from wild-type (control) animals or upon RNAi-driven btl knockdown (btl−IR) within ISCs/EBs. Dotted boxes show a magnified view of the ISCs/EBs pointed by arrows. Scale bars: 50 µm (main figure); 40 µm (close up view). e, Quantification of pERK staining intensity relative to background within ISCs/EBs in posterior midguts as in (d). Two-way ANOVA followed by Sidak’s multiple comparisons test (****P < 0.0001); n = number of ISCs/EBs from 11 posterior midguts per condition, indicated in panel. Box plots represent maxima, minima and mean. Mean value is indicated on top of boxes. Otherwise, values represent mean ± s.e.m.
Extended Data Fig. 7 Genome wide RNA pol II binding profile within adult TTC by targeted DamID (TaDa).
a, Scatterplots indicating correlation between TaDa replicates for each of the conditions used in this study. Significant correlation is observed between replicates of each condition. Each data point represents the average score for each gene (log2 Dam-pol II/ Dam-only). b, RNA Pol II binding profile to bnl in TTCs from Sucrose and Pe treated midguts. Boxes highlight areas with most prominent differences in RNA Pol II binding.
Supplementary information
Supplementary Tables 1–6
Supplementary Table 1. List of RNA Pol II-enriched transcripts in control (sucrose) and regenerating (Pe-treated) midguts identified by TaDa. UAS-Dam-RNA Pol II or UAS-Dam transgenes were expressed in adult TTCs—dSRFts>Dam-RNA Pol II (experimental) and dSRFts>Dam (normalization control)—for tracheal-specific profiling of RNA Pol II binding. The transcribed genes were annotated for Pol II binding data using a custom Perl scrip. Monte Carlo simulations were performed using randomized data to generate the FDR. A log2-transformed ratio of Dam-RNA Pol II/Dam read counts was calculated. A threshold of 1% FDR and log2 ratio >2 deems a transcript significantly bound by RNA Pol II. We identified 1,747 and 1,712 genes that were significantly bound by RNA Pol II in control (sucrose) and Pe-infected midguts, respectively. Candidate genes with functions in TTCs characterized in this study (bnl, Imp and trh) are highlighted. Supplementary Table 2. Gene ontology of RNA Pol II-enriched transcripts in control midguts. GO-term analysis of RNA Pol II-enriched transcripts in sucrose-treated midguts was performed using the ‘enrichGo’ function of the R package ClusterProfiler to search for enriched GO terms. Supplementary Table 3. Tracheal genes identified by TaDa. List of genes belonging to the ‘Epithelial tube morphogenesis’, ‘Respiratory system’ and ‘Open tracheal system’ categories of the GO-term analysis were extracted from Supplementary Table 2. Examples of genes in each of these categories that have been previously characterized for their involvement in tracheal biology are highlighted. Supplementary Table 4. RT–qPCR primers. List of RT–qPCR primers used in this study. Supplementary Table 5. List of resources used in this study. List of antibodies, chemicals, fly strains and software. Supplementary Table 6. Full list of genotypes. Description of all genotypes used in this study as they appear in each figure panel.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Perochon, J., Yu, Y., Aughey, G.N. et al. Dynamic adult tracheal plasticity drives stem cell adaptation to changes in intestinal homeostasis in Drosophila. Nat Cell Biol 23, 485–496 (2021). https://doi.org/10.1038/s41556-021-00676-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00676-z
This article is cited by
-
Developmental genetic underpinnings of a symbiosis-associated organ in the fungus-farming ambrosia beetle Euwallacea validus
Scientific Reports (2023)
-
Tracheal remodelling supports stem cells
Nature Cell Biology (2021)