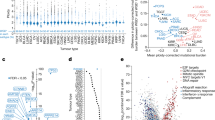
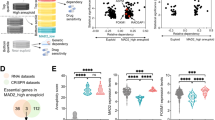
Abstract
Rewiring of cellular programmes in malignant cells generates cancer-specific vulnerabilities. Here, using an unbiased screening strategy aimed at identifying non-essential genes required by tumour cells to sustain unlimited proliferative capacity, we identify the male-specific lethal (MSL) acetyltransferase complex as a vulnerability of genetically unstable cancers. We find that disruption of the MSL complex and consequent loss of the associated H4K16ac mark do not substantially alter transcriptional programmes but compromise chromosome integrity and promote chromosomal instability (CIN) that progressively exhausts the proliferative potential of cancer cells through a p53-independent mechanism. This effect is dependent on pre-existing genomic instability, and normal cells are insensitive to MSL disruption. Using cell- and patient-derived xenografts from multiple cancer types, we show that excessive CIN induced by MSL disruption inhibits tumour maintenance. Our findings suggest that targeting MSL may be a valuable means to increase CIN beyond the level tolerated by cancer cells without inducing severe adverse effects in normal tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The accompanying RNA-seq and DNA-seq dataset is available through GEO: GSE144019. Gene sets used for GSEA are available through the MSigDB database v.7.2 at https://www.gsea-msigdb.org/gsea/msigdb/index.jsp. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Mair, B., Moffat, J., Boone, C. & Andrews, B. J. Genetic interaction networks in cancer cells. Curr. Opin. Genet Dev. 54, 64–72 (2019).
Witkiewicz, A. K. et al. Targeting the vulnerability of RB tumor suppressor loss in triple-negative breast cancer. Cell Rep. 22, 1185–1199 (2018).
Kumar, M. S. et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 149, 642–655 (2012).
Li, L. et al. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl Med. 11, eaaw7852 (2019).
He, S., Nakada, D. & Morrison, S. J. Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol. 25, 377–406 (2009).
Dick, J. E. Stem cell concepts renew cancer research. Blood 112, 4793–4807 (2008).
Hayflick, L. Mortality and immortality at the cellular level. A review. Biochemistry (Mosc.) 62, 1180–1190 (1997).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Dawson, M. A. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 (2017).
Wainwright, E. N. & Scaffidi, P. Epigenetics and cancer stem cells: unleashing, hijacking, and restricting cellular plasticity. Trends Cancer 3, 15 (2017).
Komlodi-Pasztor, E., Sackett, D., Wilkerson, J. & Fojo, T. Mitosis is not a key target of microtubule agents in patient tumors. Nat. Rev. Clin. Oncol. 8, 244–250 (2011).
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Barabe, F., Kennedy, J. A., Hope, K. J. & Dick, J. E. Modeling the initiation and progression of human acute leukemia in mice. Science 316, 600–604 (2007).
Lan, X. et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549, 227–232 (2017).
Scaffidi, P. & Misteli, T. In vitro generation of human cells with cancer stem cell properties. Nat. Cell Biol. 13, 1051–1061 (2011).
Torres, C. M. et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 353, aaf1644 (2016).
Mortimer, T. et al. Redistribution of EZH2 promotes malignant phenotypes by rewiring developmental programmes. EMBO Rep. 20, e48155 (2019).
Morales Torres, C. et al. Selective inhibition of cancer cell self-renewal through a quisinostat–histone H1.0 axis. Nat. Commun. 11, 1792 (2020).
Menyhart, O. et al. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim. Biophys. Acta 1866, 300–319 (2016).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR–Cas9 system. Science 343, 80–84 (2014).
Keller, C. I. & Akhtar, A. The MSL complex: juggling RNA–protein interactions for dosage compensation and beyond. Curr. Opin. Genet. Dev. 31, 1–11 (2015).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Galupa, R. & Heard, E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566 (2018).
Gupta, A. et al. Involvement of human MOF in ATM function. Mol. Cell Biol. 25, 5292–5305 (2005).
Gupta, A. et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol. Cell Biol. 28, 397–409 (2008).
McDonald, O. G. et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49, 367–376 (2017).
Zhu, L. et al. Expression of hMOF, but not HDAC4, is responsible for the global histone H4K16 acetylation in gastric carcinoma. Int. J. Oncol. 46, 2535–2545 (2015).
Ravens, S. et al. Mof-associated complexes have overlapping and unique roles in regulating pluripotency in embryonic stem cells and during differentiation. eLife 3, e02104 (2014).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Tonnessen-Murray, C. A., Lozano, A. & Jackson, J. G. The regulation of cellular functions by the p53 protein: cellular senescence. Cold Spring Harb. Perspect. Med. 7, a026112 (2017).
Sammons, M. A., Zhu, J., Drake, A. M. & Berger, S. L. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 25, 179–188 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Potapova, T. A., Zhu, J. & Li, R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 32, 377–389 (2013).
Lee, H. S. et al. A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC Cancer 13, 252 (2013).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Wilhelm, T., Said, M. & Naim, V. DNA replication stress and chromosomal instability: dangerous liaisons. Genes (Basel) 11, 642 (2020).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Thompson, S. L., Bakhoum, S. F. & Compton, D. A. Mechanisms of chromosomal instability. Curr. Biol. 20, R285–R295 (2010).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013).
Umbreit, N. T. et al. Mechanisms generating cancer genome complexity from a single cell division error. Science 368, eaba0712 (2020).
Tang, Y. C. & Amon, A. Gene copy-number alterations: a cost–benefit analysis. Cell 152, 394–405 (2013).
Zhu, J., Tsai, H.-J., Gordon, M. R. & Li, R. Cellular stress associated with aneuploidy. Dev. Cell 44, 420–431 (2018).
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 15, 139–150 (2018).
Santaguida, S., Tighe, A., D’Alise, A. M., Taylor, S. S. & Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 (2010).
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J. & Clarke, M. F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 100, 3983–3988 (2003).
Janssen, A., Kops, G. J. & Medema, R. H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. USA 106, 19108–19113 (2009).
Tardif, K. D. et al. Characterization of the cellular and antitumor effects of MPI-0479605, a small-molecule inhibitor of the mitotic kinase Mps1. Mol. Cancer Ther. 10, 2267–2275 (2011).
Martinez, R. et al. Mitotic checkpoint kinase Mps1 has a role in normal physiology which impacts clinical utility. PLoS ONE 10, e0138616 (2015).
Wang, Q. et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood 103, 1278–1285 (2004).
Kusakabe, K. et al. Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity. J. Med. Chem. 58, 1760–1775 (2015).
Kim, D. et al. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat. Struct. Mol. Biol. 17, 1027–1029 (2010).
Kadlec, J. et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat. Struct. Mol. Biol. 18, 142–149 (2011).
Baell, J. B. et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257 (2018).
MacPherson, L. et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature 577, 266–270 (2020).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: genome-Wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 12, e1004873 (2016).
Henser-Brownhill, T., Monserrat, J. & Scaffidi, P. Generation of an arrayed CRISPR–Cas9 library targeting epigenetic regulators: from high-content screens to in vivo assays. Epigenetics 12, 1065–1075 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J 17, 10–12 (2011).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Karolchik, D. et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Scheinin, I. et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 24, 2022–2032 (2014).
Kyriacou, E. & Heun, P. High-resolution mapping of centromeric protein association using APEX-chromatin fibers. Epigenetics Chromatin 11, 68 (2018).
Bui, M. et al. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150, 317–326 (2012).
Acknowledgements
We thank the Crick Biological Research Facility for help with the animal work, the Crick Advanced Sequencing for preparing and sequencing NGS libraries, the Crick Light Microscopy group for help with fluorescence microscopy, and the Flow Cytometry group for help with cell sorting. We thank T. Metz and Oncotest–Charles River for sharing PDX lines and P. Zalmas, A. Tedeschi and E. Gronroos for useful discussions and sharing reagents. This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001152), the UK Medical Research Council (FC001152), and the Wellcome Trust (FC001152), and by the CRUK Drug Discovery Award (C50796/A19448) provided by Cancer Research UK and Bayer Healthcare. Y.D. and M.B. were funded by the Intramural Research Program of the National Institutes of Health, USA.
Author information
Authors and Affiliations
Contributions
J.M. performed most of the experiments and analyses. C.M.T. and P.S. helped with in vivo experiments and microscopy. L.R., T.S.W. and M.J. generated the Cas9-expressing cell lines. H.P. performed NGS analysis of the CRISPR-KO screens and RNA-seq. M.-C.D. performed scanning electron microscopy. S.H. performed karyotyping analysis. O.R.S. performed high-content microscopy analysis. M.C. performed low-pass genome sequencing. M.B. and Y.D. analysed chromatin fibres. P.S. conceived and supervised the study and wrote the manuscript with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Mark Dawson, Marcel van Vugt and the other, anonymous, reviewer for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Generation of a reporter cell line to assess loss of unlimited proliferative capacity.
a, Schematics of the experimental system used for the screen. b, Diagram illustrating the design of the DCN-driven fluorescent reporter. Arrows: approximate location of primers used in c, ORF: open reading frame. c, Amplicons detecting the correctly integrated IRES-GFP cassette within the DCN 3’UTR. Edited: TDF reporter line; unedited: unedited parental cells. Schematics at the top show the sequence junctions probed. MW: molecular weight. d, Quantification of DCN upregulation in SSEA1− non-tumorigenic tumor cells as assessed by microarray analysis16. Values are average ± SEM from n = 3 biologically independent tumors. P-value from one-tailed Student’s t-test. e, Flow cytometry analysis of untreated or quisinostat-treated, differentiated TDF reporter cells. f, Flow cytometry analysis of a tumor containing the DCN-GFP reporter and of a control tumor induced by unedited TDF cells. Gates used for sorting cells used in h. g, Quantification of DCN upregulation in SSEA1− non-tumorigenic tumor cells, as assessed by flow cytometry (FACS) detecting expression of the DCN-GFP reporter. Values are average ± SEM from n = 5 biologically independent tumors. P-value from one-tailed Student’s t-test. h, Transplantation assays for secondary tumor formation. Quantification of tumors induced by injection of 1,000 SSEA1+ or GFP+ sorted cells. N = 4 independent injections per condition. P-value from one-sided Fisher’s exact test. i, Quantification of DCN upregulation in HRAS-KO tumors as assessed by flow cytometry detecting expression of the DCN-GFP reporter. Individual values from two biologically independent samples. P-value from one-tailed Student’s t-test. j, Clonogenic assays showing loss of unlimited proliferative capacity by HRAS-KO TDF cells. Individual values from 2 biologically independent samples. P-value from one-tailed Student’s t-test. Scale bar: 100 µm. k, Growth kinetics of tumors induced by wild-type (WT) or HRAS-KO TDF cells. Values are average ± SEM from n = 4 biologically independent tumors. P-value from one-tailed Student’s t-test at the last time point. l, Growth kinetics of WT or HRAS-KO TDF cells as assessed by MTS assays. Values normalized to initial plating show the average ± SEM from n = 3 biologically independent samples. P-value from one-tailed Student’s t-test at the last time point. Source data are provided.
Extended Data Fig. 2 CRISPR-Cas9 screen controls.
a, Relative abundance of sgRNAs in the two biological replicates of TDF cells transduced with the sgRNA library 14 d after Cas9 induction (T14). NTC: non-targeting sgRNA controls. The statistical significance of the correlation between the two replicates (two-sided Spearman correlation test) and the corresponding correlation coefficient (rs) are indicated. b, Fold change (FC) in sgRNA abundance comparing the T14 and T0 cell populations of replicate 2 (Rep 2). Individual, alphabetically ordered sgRNAs are shown. Non-targeting sgRNA controls (NTC) are spread among targeting sgRNAs rather than clustered to ease their visualization. c, Proportion of ribosomal genes depleted (average log2 FC ≤ -0.5) at T14, indicating near saturation of the screen. Four out of 83 ribosomal genes targeted in the library are not expressed in TDF cells and are excluded from the analysis. The average value from both replicates is shown. d, Fold change in sgRNA abundance comparing the T24 and T0 cell populations in the counter-screen (see Methods). The average (left) or cumulative (right) value of all sgRNAs targeting each gene is plotted. The average of both replicates is shown in the graph on the left. The graph on the right displays only depleted genes and omits ribosomal genes due to the large number of sgRNAs targeting each gene (1-42 sgRNAs, median: 14 sgRNAs/gene), which makes them not comparable with other targeted genes. The essential gene POLR2A is shown as a positive control of sgRNA depletion. NTC: non-targeting sgRNA controls. e, Filtering strategy used to select hits from both screen arms. Source data are provided.
Extended Data Fig. 3 In vivo validation and characterization of primary hits.
a, Growth kinetics of the indicated conditions in tumor maintenance assays performed as indicated in Fig. 1e. N = 4 biologically independent tumors. P-value from one-tailed Student’s t-test at the last time point. b, Quantification of gene editing efficiency of TDF cells of the indicated samples as assessed by Sanger sequencing and subsequent TIDE analysis https://tide.deskgen.com/52. Cntr: control cells expressing GFP-targeting sgRNAs. c, Schematic representation of the four MSL subunits and the mutants generated by genome editing. Arrowheads indicate the approximate location of sgRNAs used to generate truncated proteins lacking functional domains used in d, The characterized function of each domain is described in Supplementary Table 3. CC: coiled-coil. CD: chromo. CB: chromobarrel. d, Quantification of MSL loss-of-function in the indicated MSL-mutant cell populations generated using the sgRNAs indicated in c, H4K16ac levels were measured by quantitative immunofluorescence and used as a readout of MSL function. The effect of KAT8-KO is underestimated as many cells H4K16aclow died and detached from the plate prior to staining. Note that slight differences in the fraction of H4K16aclow cells across mutants may be due to differences in sgRNA activity. All mutants resulted in reduction of the histone mark. The number of analyzed cells in each condition is shown. The statistical significance of the differences compared to a control sgRNA are indicated by three asterisks (p < 0.0001, two-tailed Fisher’s test) e-f, Representative images (e) and quantification (f) of H4K16ac levels in TDF cells at the indicated times after induction of MSL1 knock-out. Values are average ± SEM from n = 3 biologically independent samples. P-value from one-way ANOVA. Scale bar: 20 µm. g, Quantification of gene editing in the indicated tumors by Sanger sequencing and subsequent TIDE analysis. Tumors showing less than 50% edited sequence were considered unedited. Note the varying editing efficiency across all tumors, indicating the presence of wild-type cells also in tumors classified as “edited”. Wild-type cells sustain tumor growth, leading to an underestimation of the effect induced by MSL disruption. Source data are provided.
Extended Data Fig. 4 Disruption of the MSL complex impairs the long-term proliferative capacity of cancer cells.
a, Quantification of the number of adherent, viable cells 8 d after induction of KAT8 knock-out with doxycycline (KAT8-KO) and in the corresponding uninduced cells. Values are average ± SEM from n = 3 biologically independent samples. P-value from one-tailed Student’s t-test. b, Images of TDF cells 4 d after transfection of synthetic crRNAs-trRNAs targeting KAT8 (KAT8-KO) or the GFP reporter (cntr-KO). Scale bar: 1 mm. c, Quantification of H4K16ac levels in adherent, alive cells and detached cells recovered from the medium 4 d after induction of KAT8 knock-out by doxycycline treatment. For boxplots, top, middle and bottom delimiters: 75th, 50th, 25th percentiles; top and bottom whiskers: 90th and 10th percentiles. N = 471 and 488 biologically independent adherent and detached cells, respectively. P-value from one-tailed Mann Whitney’s U-test. AU: arbitrary units. d, Limiting dilution clonogenic assays of the indicated TDF cells 11 d after plating. Representative images of 96-well plates in which 2-fold serial dilutions of cells were plated. External wells are not shown (see Methods). Yellow shows the IncuCyte cell mask detecting growing clones. Scale bar: 6 mm. e, Limiting dilution clonogenic assays using TDF cells. The percentage of wells containing at least one clone larger than 20 cells is plotted. Values are average ± SEM from n = 3 biologically independent samples. The knock-out efficiency, as assessed by quantification of H4K16aclow cells in each population is indicated. P-value from one-tailed Student’s t-test. f, Limiting dilution transplantation assay into NSG mice using cells from unedited or MSLLOF (MSL3-KO) tumors. The frequency of tumor-propagating cells (estimate with upper and lower limits) is indicated. P-value from χ2 test. g-h, Quantification (g) and representative images (h) of H4K16ac levels in HT-1080 cells at the indicated times after induction of MSL1 knock-out. Values are average ± SEM from n = 3 areas in the well for a total of over 800 cells per condition. P-value from two-tailed Fisher’s exact test. Scale bar: 100 µm. i, Viability phenotype of the indicated knock-out mice. IMPC: International Mouse Phenotyping Consortium (see: https://www.mousephenotype.org/data/genes/MGI:1341851). PMID: Pubmed ID. Source data are provided.
Extended Data Fig. 5 Gene expression and genomic abnormalities in MSL-disrupted cells.
a, Karyotyping of MSLLOF (MSL3-KO) TDF cells. The lines represent the consensus value for genomic segments that are either gained (positive values) or lost (negative values) relative to the rest of the genome (see Methods). The black dots are experimental values for 1 Mb intervals. X and Y chromosomes, which affects the performance of QDNASeq66, are excluded from the analysis. b, Correlation between large-scale changes in mRNA levels and DNA copy number alterations in MSLLOF (MSL3-KO) cells. For each positional gene set identified as upregulated or downregulated in MSLLOF cells by GSEA analysis, the relative copy number changes detected by CNVkit are shown. Chromosome 1 is shown to illustrate that even though CNVKit did not call copy number changes, likely because only a subset of cells in the population were affected by consistent gains or losses, the p arm showed a relative reduction compared to control cells (cntr-KO), correlating with apparent downregulation of the corresponding genes. The slight difference compared to the Chr.1 pattern observed in the comparison with WT cells is likely due to subsampling of the population upon sgRNA transfection in the cntr-KO sample. Note that the genetic drivers in TDF cells are located in chromosomal regions showing highly significant changes, which indicates high prevalence in the population of MSLLOF cells: HRAS on chr11p15 - gained, p53 on chr17p13 – lost, and hTERT on chr5 p15 – gained. This pattern suggests selection of less deleterious karyotypes in the population, compared to random gains or losses of other regions.
Extended Data Fig. 6 Accumulation of single-stranded DNA upon MSL disruption.
a-g, Immunofluorescence microscopy of MSLLOF TDF cells using the indicated antibodies (a-d, f-g) and quantification of DAPI intensity in nuclei and micronuclei (e). Nuclei were counterstained with DAPI. Yellow arrowheads in g indicate mitotic cells displaying p-RPA foci on chromosomes. N = 172 biologically independent cells collected from two experiments. P-value from one tailed paired Student’s t-test. Scale bar: 10 µm. h, Aphidicolin sensitivity assay comparing TDF and normal HME1 cells. Values are average ± SEM from n = 3 biologically independent samples after 5 days of growth. The non-linear fit of the experimental points is shown. Minor differences in TDF IC50 compared to Fig. 4g are due to a slightly different set up of the experiment (see Methods). i, Clonogenic assays comparing the proliferative capacity of individual TDF cells of the indicated genotypes (See also Fig. 6a). N = 103, 131, 161 biologically independent clones for WT, cntr-KO and MSLLOF(MSL3-KO), respectively, collected from two experiments. Horizontal lines: median values. P-value from one-way ANOVA followed by Dunn’s test. N.s.: non-significant (p-value = 0.9495). Source data are provided.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Table 1: Pooled sgRNA library with negative and positive controls. Supplementary Table 2: Screen and counter-screen analysis. Supplementary Table 3: Function of protein domains of MSL subunits. Supplementary Table 4: Characteristics of the human PDX cell lines used in this study. Supplementary Table 5: IMPC mouse data analysis. Supplementary Table 6: DEGs in MSL-KO cells (MSL1-KO, MSL2-KO and MSL3-KO) compared to control cells (GFP-KO and WT). Supplementary Table 7: GSEA analysis comparing MSLLOF (MSL1-KO, MSL2-KO and MSL3-KO) and control (WT and GFP-KO) cells. Supplementary Table 8: Copy number changes in MSL3-KO TDF cells compared to WT and cntr-KO cells detected using a CNVKit. Supplementary Table 9: Cell line growth and transfection conditions. Supplementary Table 10: sgRNA sequences cloned into pLX- or pLenti-sgRNA plasmids and used in the study. Supplementary Table 11: List of primers used in this study. Supplementary Table 12: Analysis steps for automated quantification of pRPA foci using Harmony software.
Supplementary Video 1
Live imaging of a mitotic WT TDF cell.
Supplementary Video 2
Live imaging of a mitotic MSL3-KO TDF cell.
Supplementary Video 3
Live imaging of a mitotic MSL3-KO TDF cell showing unaligned chromosomes.
Supplementary Video 4
Live imaging of a mitotic MSL3-KO TDF cell showing unaligned chromosomes.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Monserrat, J., Morales Torres, C., Richardson, L. et al. Disruption of the MSL complex inhibits tumour maintenance by exacerbating chromosomal instability. Nat Cell Biol 23, 401–412 (2021). https://doi.org/10.1038/s41556-021-00657-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00657-2
This article is cited by
-
H4K16ac activates the transcription of transposable elements and contributes to their cis-regulatory function
Nature Structural & Molecular Biology (2023)
-
Mechanisms of chromosomal instability (CIN) tolerance in aggressive tumors: surviving the genomic chaos
Chromosome Research (2023)
-
Molecular differences in renal cell carcinoma between males and females
World Journal of Urology (2023)
-
Two distinct males absent on the first (MOF)-containing histone acetyltransferases are involved in the epithelial–mesenchymal transition in different ways in human cells
Cellular and Molecular Life Sciences (2022)
-
MSL pushes genomic instability over the edge
Nature Cell Biology (2021)