Abstract
Intestinal stem cells (ISCs) are located at the crypt base and fine-tune the balance of their self-renewal and differentiation1,2, but the physiological mechanism involved in regulating that balance remains unknown. Here we describe a transcriptional regulator that preserves the stemness of ISCs by restricting their differentiation into secretory-cell lineages. Interferon regulatory factor 2 (IRF2) negatively regulates interferon signalling3, and mice completely lacking Irf24 or with a selective Irf2 deletion in their intestinal epithelial cells have significantly fewer crypt Lgr5hi ISCs than control mice. Although the integrity of intestinal epithelial cells was unimpaired at steady state in Irf2-deficient mice, regeneration of their intestinal epithelia after 5-fluorouracil-induced damage was severely impaired. Similarly, extended treatment with low-dose poly(I:C) or chronic infection of lymphocytic choriomeningitis virus clone 13 (LCMV C13)5 caused a functional decline of ISCs in wild-type mice. In contrast, massive accumulations of immature Paneth cells were found at the crypt base of Irf2−/− as well as LCMV C13-infected wild-type mice, indicating that excess interferon signalling directs ISCs towards a secretory-cell fate. Collectively, our findings indicate that regulated interferon signalling preserves ISC stemness by restricting secretory-cell differentiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The microarray datasets have been deposited in the Gene Expression Omnibus under the accession numbers GSE137189 and GSE137190. ChIP–Seq datasets have been deposited under the accession number GSE137260. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Metcalfe, C., Kljavin, N. M., Ybarra, R. & de Sauvage, F. J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159 (2014).
Harada, H. et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58, 729–739 (1989).
Matsuyama, T. et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97 (1993).
Wilson, E. B. et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013).
Pestka, S., Krause, C. D. & Walter, M. R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32 (2004).
Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 (2001).
Taniguchi, T. & Takaoka, A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2, 378–386 (2001).
Sato, T. et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 15, 696–700 (2009).
Soares, P. M. et al. Inflammatory intestinal damage induced by 5-fluorouracil requires IL-4. Cytokine 61, 46–49 (2013).
Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283 (2002).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Ireland, H. et al. Inducible cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126, 1236–1246 (2004).
Hida, S. et al. CD8+ T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-α/β signaling. Immunity 13, 643–655 (2000).
Fink, K. & Grandvaux, N. STAT2 and IRF9: beyond ISGF3. JAKSTAT 2, e27521 (2013).
Wu, X. et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 172, 423–438 (2018).
Sun, L. et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe 17, 85–97 (2015).
Rusinova, I. et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, D1040–D1046 (2013).
Subramanian, A., Kuehn, H., Gould, J., Tamayo, P. & Mesirov, J. P. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 3, 3251–3253 (2007).
King, S. L., Mohiuddin, J. J. & Dekaney, C. M. Paneth cells expand from newly created and preexisting cells during repair after doxorubicin-induced damage. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G151–G162 (2013).
Andreu, P. et al. A genetic study of the role of the Wnt/β-catenin signalling in Paneth cell differentiation. Dev. Biol. 324, 288–296 (2008).
Lee, G. et al. Contributions made by CDC25 phosphatases to proliferation of intestinal epithelial stem and progenitor cells. PLoS ONE 6, e15561 (2011).
Yan, K. S. et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 545, 238–242 (2017).
VanDussen, K. L. et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497 (2012).
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y. & Wrana, J. L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015).
Kamphuis, E., Junt, T., Waibler, Z., Forster, R. & Kalinke, U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood 108, 3253–3261 (2006).
Le Bon, A. et al. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol. 176, 2074–2078 (2006).
Yilmaz, Ö. H. et al. TORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–793 (2018).
Huang, daW., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank H. Kamioka for secretarial support; K. Shiseki, S. Kuroda, S. Yamada and M. Kajita for technical support; T. W. Mak (Campbell Family Institute for Breast Cancer Research, Princess Margaret Cancer Centre) for providing the Irf2–/– mice; D. J. Winton (Cancer Research UK Cambridge Institute) for providing the Ah-cre mice; U. Kalinke (Hanover Medical School) for providing Ifnar1-flox mice; T. Akatsuka (Saitama Medical University) for providing the LCMV C13; and M. Nakagawa (TMDU) and Y. Asahina (TMDU) for providing the patient information. This work was supported by the Naito Foundation (T.O.); a Naoki Tsuchida Research Grant (Taku Sato); a Research Grant from Mishima Kaiun Memorial Foundation (Taku Sato); Nanken-Kyoten, TMDU (Toshiro Sato); a Grant-in-Aid for Scientific Research on Innovative Areas ‘Stem Cell Aging and Disease’ from MEXT, Japan (grant nos 15H01508 and 17H05635; T.O.); and the Japan Science and Technology Agency, PREST (grant no. JPMJPR13M4; Taku Sato).
Author information
Authors and Affiliations
Contributions
Taku Sato conceived the study, performed experiments, analysed data and wrote the manuscript. S.I. performed some of the bioinformatics analysis, some of the organoid culture experiments, some of the immunostaining and analysed the data. J.A. performed some of the immunostaining and analysed the data. H.Y. analysed the TEM images, performed in situ hybridization and analysed the data. M.F. and Toshiro Sato performed organoid culture experiments and the analysis of human intestinal epithelium, and provided advice and discussion on some parts of this study. K.Y., K.K, T.A. and R.O. performed the histological examination of human colon tissues and provided advice and discussion on some parts of this study. T.O. supervised the overall project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
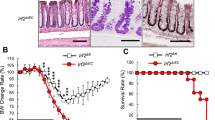
Extended Data Fig. 1 Intestinal epithelial cell profiling by flow cytometry, Irf2 expression in steady state WT mice, and apoptosis and Wnt/Notch signalling in mucositis-induced Irf2−/− mice.
a, Flow cytometry gating strategy for ISCs and Paneth cells in Lgr5-EGFP-Ires-CreERT2 mice. ISCs and Paneth cells were isolated as Lgr5hi cells and CD24hiUEA1hi cells within EpCAMhi crypt ECs, respectively. b, Representative images of Irf2 mRNA detection in the jejunum epithelium of WT mice using RNAscope in situ hybridization assays. 3 mice. Irf2 mRNA is depicted as brown dots. Most epithelial cells, including ISCs (red arrowheads), transit amplifying cells (red dashed lines) and goblet cells (Gob, black arrowheads), expressed Irf2 mRNA. Orange arrows indicate Irf2 mRNA dots in ISCs. c-e, Gene expression of Olfm4 (c), Lyz1 (d) and Irf2 (e) in FCM-sorted ISCs and in Paneth cells relative to total crypt ECs. Data represent means ± s.d. of n=3 mice. Statistics were determined by one-way ANOVA followed by Tukey’s multiple comparisons test. f, Representative images of TUNEL staining in the jejunum of Irf2+/− and of Irf2−/− mice injected with 5-FU as in Fig. 1. Intestines were harvested on d 2 and 6 (Irf2+/−, naïve, 3 mice; Irf2+/−, day 2, 3 mice; Irf2+/−, day 6, 4 mice; Irf2–/−, naïve, 3 mice; Irf2–/−, day 2, 3 mice; Irf2–/−, day 6, 4 mice). g, Gene expression of Olfm4, Ascl2 and Hes1 in the jejunum of Irf2−/− mice compared with Irf2+/− mice on day 6 after 5-FU injection. Data represent means ± s.d. of n=4 mice. Statistics were determined by two-tailed Mann-Whitney test. Scale bars: 20 μm (b) and 100 μm (f).
Extended Data Fig. 2 Generation of an Irf2 conditional-KO mouse.
a, Schematic representation of the targeting vector and expected gene replacement at the Irf2 locus. Blue triangles, loxP sequences; green triangles, FRT sequences. b, The floxed allele was genotyped by detecting the 3′-loxP sequence by PCR. Data are representative of two independent experiments with similar results. WT allele, 332 bp; floxed Irf2 allele, 376 bp. c, Long-range genomic PCR analysis of the floxed-out allele in tissues isolated from Irf2ΔIEC mice. Data are representative of two independent experiments with similar results. WT allele, 3 kbp; KO allele, 880 bp. d, Relative Irf2 expression in the spleen, whole jejunum and isolated crypts of Irf2ΔIEC (n = 3) compared with Irf2fl/fl (n = 3) mice. Data represent means ± s.d. Statistics were determined by one-way ANOVA followed by two-tailed Dunnett’s multiple comparisons test. e, Naïve Irf2ΔIEC mouse jejunum stained with H&E. Representative image from five mice. The crypt-villus structure of Irf2ΔIEC mice was normal in the steady state. Scale bar: 300 μm.
Extended Data Fig. 3 Short-term lineage tracing of ISCs in Irf2–/– mice, and ISC reduction in Irf2ΔAh-Cre; Lgr5ki mice.
a, Lineage tracing in ISCs from Irf2–/–;Lgr5ki;R26R or Irf2+/−;Lgr5ki;R26R mice. Jejunum was harvested 72 hrs after tamoxifen injection and stained with X-gal. Image representative of four mice per group. b, The relative migration distance of Lgr5+ ISC-derived progenies was calculated as (Traced X-gal+ cell height)/(total crypt-villus height) x 100 (%) in each crypt-villus unit. Each dot (n) represents the relative migration distance of one individual among the Lgr5+ ISC-derived progenies. Data represents mean relative migration distances ± s.d. of n=200 Lgr5+ ISC-derived progenies from four mice each. c-e, Representative FCM profiles (c), number of Lgr5+ ISCs (d), and organoid-forming efficiency (e) of β-naphthoflavone (βNF)-treated Irf2ΔAh-Cre; Lgr5ki mice and in control Lgr5ki mice (n = 5 mice per group in c, d, n = 4 experiments per group in e). Data represent means ± s.d. f,g, Jejunum sections of 5-FU treated Irf2fl/fl and Irf2ΔAh-Cre mice at one month after β-NF treatment, stained with Ki67, representative of five mice per group (f). Viable regenerating crypts per intestinal circumference were counted in the Ki67-stained jejunum from these mice (Irf2fl/fl mice: n = 5 mice, average number of viable crypts in 5–7 sections per mouse were counted, Irf2ΔAh-Cre mice: n = 5 mice, average number of viable crypts in 5–7 sections per mouse were counted) (g). Data represent means ± s.d. Statistics were determined by two-tailed Mann-Whitney test. Scale bars: 300 μm (a) and 500 μm (f).
Extended Data Fig. 4 Attenuated crypt regeneration capacity in Irf2–/–; Ifnar1–/– and Irf2–/–; Ifnar1–/–; Stat1–/– mice, and the expression of IFN-inducible genes in the Irf2–/–; Ifnar1–/–; Stat1–/– mice.
a, Jejunum sections from 5-FU-treated Irf2+/–;Ifnar1–/– and Irf2–/–;Ifnar1–/– mice were stained with Ki67. Image representative of five mice per group. Scale bars: 500 µm. b, Viable regenerating crypts per intestinal circumference were counted in the Ki67-stained jejunum from Irf2+/−;Ifnar1–/– (n = 6 mice, average number of viable crypts of 5–7 sections per mouse were counted) and from Irf2−/−;Ifnar1–/– (n = 5 mice, average number of viable crypts of 5–7 sections per mouse were counted) mice. Data represents means ± s.d. c, Jejunum sections from 5-FU-treated Irf2+/−;Ifnar1–/–;Stat1–/– and Irf2−/−;Ifnar1–/–;Stat1–/– mice were stained with Ki67. Image representative of five mice per group. Scale bars: 500 µm. d, Viable regenerating crypts per intestinal circumference were counted in the Ki67-stained jejunum from Irf2+/−;Ifnar1–/–;Stat1–/– (n = 5 mice, average number of viable crypts of 5–7 sections per mouse were counted) and from Irf2−/−;Ifnar1–/–;Stat1–/– (n = 5 mice, average number of viable crypts of 5–7 sections per mouse were counted) mice. Data represent means ± s.d. e-g, The expression of Ifit1 (e), Ifit2 (f) and Tapbp (g) in isolated crypt epithelial cells of Irf2+/– and of Irf2–/– mice (n = 5 or 7 mice per group). h-j, The expression of Ifit1 (h), Ifit2 (i), Tapbp (j) in FCM sorted crypt ECs of Irf2+/–;Ifnar1–/–;Stat1–/– mice and of Irf2–/–;Ifnar1–/–;Stat1–/– mice (n = 5 or 7 mice per group). Data represent means of relative expression ± s.d. Statistics were determined by two-tailed Mann-Whitney test.
Extended Data Fig. 5 Chronic viral infection reduces stem-cell function in ISCs.
a,b Ifit1 mRNA expression (a) and organoid-forming ability of FCM-sorted crypt epithelial cells (b) from uninfected and from 7 d post-infection of LCMV C13-infected WT mice. Data represent means ± s.d. of n=4 mice in (a), and n=6 mice in (b). Statistics were determined by two-tailed Mann-Whitney test. (c) Ileal sections from uninfected (5 mice, upper) and from LCMV C13 infected (7 mice, lower) WT mice stained with Alcian Blue. Right panel shows higher magnification of the indicated region. Scale bars: 200 μm (lower magnification images in c) and 20 μm (higher magnification images in c).
Extended Data Fig. 6 Direct IFN signalling reduces organoid formation both in mouse and human intestinal epithelial cells.
Organoid-forming abilities of mouse crypts (a,b) and human small intestinal epithelial cells (c,d) in the presence or absence of IFNα were evaluated. a, Mouse IFNα was added to the crypt culture of mouse small intestine from d 0 to 3, then PI staining was performed and images were obtained at day 3 (upper panels), or IFNα was added from d 3 to 6, then PI staining was performed and images were obtained at day 6 (lower panels). Images are representative of two independent experiments with similar results. b, Organoid-forming efficiency of mouse crypts in the presence or absence of IFNα was evaluated after six d of culture. Data represent mean efficiency ± s.d. of n=5 mice. c, Human IFNα2a was added to the organoid culture of human intestinal epithelial cells derived from two different donors (Clone#1, Clone#2) from d 0 to 6, then organoid images were obtained at day 6. Images are representative of four independent experiments with similar results. d, Organoid-forming efficiency of human small intestinal epithelial cells in the presence or absence of IFNα was evaluated after six d of culture. Data represent mean efficiencies ± s.d. of n=4 independent experiments. Statistics were determined by one-way ANOVA followed by two-tailed Dunnett’s multiple comparisons test. Scale bars: 500 μm (a) and 1 mm (c).
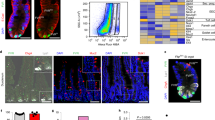
Extended Data Fig. 7 Microarray analyses of ISCs from poly(I:C)-treated Ifnar1−/−; Lgr5ki versus Lgr5ki mice, and those from β-naphthoflavone-treated Irf2ΔAh-Cre; Lgr5ki versus Irf2fl/fl; Lgr5ki mice.
a, Heat map showing differentially expressed genes (DEGs) in ISCs from poly(I:C)-treated Ifnar1−/−;Lgr5ki mice and Lgr5ki mice (Lgr5ki, n = 2 replicates, 3 or 4 mice pooled per replicate, Ifnar1-/-;Lgr5ki, n = 2 replicates, 3 mice pooled per replicate). b, Heat map showing DEGs in ISCs of β-naphthoflavone (βNF)-treated Irf2ΔAh-Cre; Lgr5ki mice and Irf2fl/fl;Lgr5ki mice (n = 4 replicates from 4 mice per group).
Extended Data Fig. 8 Secretory cell hyperplasia in a patient with HCV who received IFN therapy.
Images of alcian blue and H&E staining of colon tissues from an HCV uninfected patient (upper), from an HCV infected patient who had not received IFN therapy (middle) and from an HCV infected patient who had received IFN therapy (lower, 2 months after 11 months of IFN therapy completion). Scale bars: 100 μm.
Extended Data Fig. 9 Paneth cell abnormalities in Irf2-deficient mice.
a, Crypt ECs of Irf2+/− mice were subdivided into four fractions based on CD24 and UEA1 expression; CD24hiUEA1hi mature Paneth cells, CD24hiUEA1lo cells, CD24intUEA1lo cells or CD24loUEA1neg cells. The cells were FCM sorted and stained with PAS, a marker for secretory-lineage cells. The FACS profile and images are representative of two independent experiments. b, FCM-sorted UEA1hi mature Paneth cells and UEAlo immature Paneth cells from Irf2fl/fl mice and from Irf2ΔIEC mice observed using transmission electron microscopy (TEM). Representative TEM images of each cell population are shown (20 cells per cell fraction). Scale bars: 10 μm (a) and 1 μm (b). c, Granule size of each cell population was measured. Each dot represents the average granule size of one individual cell. Data represent means ± s.d. of n=20 cells in each cell fraction. Statistics were determined by one-way ANOVA followed by Tukey‘s multiple comparisons test. d,e Representative FACS profiles (gated on EpCAMhi crypt ECs) (d) and number (e) of mature Paneth cells (CD24hiUEA1hi) in Irf2+/– and in Irf2–/– mice (n = 5 mice per group). Data represent means ± s.d. Statistics were determined by two-tailed Mann-Whitney test.
Extended Data Fig. 10 Altered YAP, Wnt and Notch signalling in chronically IFN-stimulated ISCs, and IRF2-binding loci detected in ISCs.
a-f, GSEA for ISCs from βNF-treated Irf2ΔAh-Cre; Lgr5ki (blue) and from control Lgr5ki (red) mice (a-c, n = 4 replicates from 4 mice per group) or from poly(I:C)-treated Lgr5ki (blue) mice and Ifnar1−/−;Lgr5ki (red) mice (d-f, Lgr5ki, n = 2 replicates, 3 or 4 mice pooled per replicate, Ifnar1-/-;Lgr5ki, n = 2 replicates, 3 mice pooled per replicate) using gene-set signatures for the YAP signature (a,d), Wnt signature (YAP suppressed genes, b,e) and Notch targets (c,f). NES, normalized enrichment score. g-j, IRF2 Chip-seq profiles of the loci of well-known IFN-inducible genes (Parp12 (g), Nmi (h) and Tap2 (i)) in ISCs. Input represents the sequence profile of genomic DNA used in CHIP pull-downs. Red arrows indicate IRF2-binding peaks. Top-ranking known motifs within the IRF2-binding peaks in ISCs were identified with HOMER (see Methods) (j). Data are representative of two independent experiments with similar results. k, Venn diagram showing the overlap of IRF2-bound targets (IRF2 ChIP-seq), which was also experimentally validated ISGs in the Interferome database, and genes (Microarray) that were upregulated >1.5-fold in ISCs of polyI:C-treated WT mice versus Ifnar1−/− mice.
Supplementary information
Supplementary Tables 1–12
Supplementary Table 1. GOTERM_BP ‘defence response to virus’ genes. Related to Fig. 4a, genes included in GOTERM BP ‘defence response to virus’ are shown. Red text reveals IFN-inducible genes defined by INTERFEROME v2.0 database. DEGs (n = 781) upregulatred in ISCs of poly(I:C)-treated Lgr5ki mice compared with those of similarly treated Ifnar1−/−; Lgr5ki mice, were applied to GO analysis using the DAVID tool and calculated uncorrected P value. Supplementary Table 2. GOTERM CC ‘extracellular region’ genes. Related to Fig. 4b, genes included in GOTERM CC ‘extracellular region’ genes are shown. Red text reveals genes encoding antibacterial proteins and endocrine hormones. DEGs (n = 781) upregulatred in ISCs of poly(I:C)-treated Lgr5ki mice compared with those of similarly treated Ifnar1−/−; Lgr5ki mice, were applied to GO analysis using the DAVID tool and calculated uncorrected P value. Supplementary Table 3. ISC signature. Supplementary Table 4. Progenitor signature. Supplementary Table 5. Paneth cell signature (GSE25109). Supplementary Table 6. Secretory Progenitor signature (GSE51398). Supplementary Table 7. Enterocyte Progenitor signature (GSE51398). Supplementary Table 8. YAP signature25. Supplementary Table 9. Wnt signature25. Supplementary Table 10. Notch signature (GSE66751). Supplementary Table 11. Summary of primers used in this study. Supplementary Table 12. Summary of antibodies used in this study.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 2
Unprocessed gel image data
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 9
Statistical source data
Rights and permissions
About this article
Cite this article
Sato, T., Ishikawa, S., Asano, J. et al. Regulated IFN signalling preserves the stemness of intestinal stem cells by restricting differentiation into secretory-cell lineages. Nat Cell Biol 22, 919–926 (2020). https://doi.org/10.1038/s41556-020-0545-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0545-5
This article is cited by
-
Advanced Progression for the Heterogeneity and Homeostasis of Intestinal Stem Cells
Stem Cell Reviews and Reports (2023)
-
Challenges and opportunities targeting mechanisms of epithelial injury and recovery in acute intestinal graft-versus-host disease
Mucosal Immunology (2022)
-
Differential regulation of β-catenin-mediated transcription via N- and C-terminal co-factors governs identity of murine intestinal epithelial stem cells
Nature Communications (2021)
-
Tissue regulatory T cells: regulatory chameleons
Nature Reviews Immunology (2021)
-
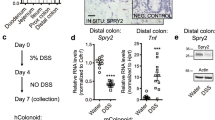
IRF2 maintains the stemness of colonic stem cells by limiting physiological stress from interferon
Scientific Reports (2020)