Abstract
The linear ubiquitin chain assembly complex (LUBAC), which consists of HOIP, SHARPIN and HOIL-1L, promotes NF-κB activation and protects against cell death by generating linear ubiquitin chains. LUBAC contains two RING-IBR-RING (RBR) ubiquitin ligases (E3), and the HOIP RBR is responsible for catalysing linear ubiquitination. We found that HOIL-1L RBR plays a crucial role in regulating LUBAC. HOIL-1L RBR conjugates monoubiquitin onto all LUBAC subunits, followed by HOIP-mediated conjugation of linear chains onto monoubiquitin, and these linear chains attenuate the functions of LUBAC. The introduction of E3-defective HOIL-1L mutants into cells augmented linear ubiquitination, which protected the cells against Salmonella infection and cured dermatitis caused by reduced LUBAC levels due to SHARPIN loss. Our results reveal a regulatory mode of E3 ligases in which the accessory E3 in LUBAC downregulates the main E3 by providing preferred substrates for autolinear ubiquitination. Thus, inhibition of HOIL-1L E3 represents a promising strategy for treating severe infections or immunodeficiency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Source data for Figs. 1–8 and Extended Data Figs. 1–5, 7 and 8 have been provided as statistical source data or unprocessed blots. MS raw data have been deposited in the ProteomeXchange database with the accession code PXD018038. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Mukhopadhyay, D. & Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 (2007).
Wickliffe, K., Williamson, A., Jin, L. & Rape, M. The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 109, 1537–1548 (2009).
Zinngrebe, J., Montinaro, A., Peltzer, N. & Walczak, H. Ubiquitin in the immune system. EMBO Rep. 15, 28–45 (2014).
Kornitzer, D. & Ciechanover, A. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol. 182, 1–11 (2000).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
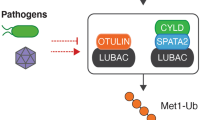
Iwai, K., Fujita, H. & Sasaki, Y. Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 (2014).
Sasaki, K. & Iwai, K. Roles of linear ubiquitinylation, a crucial regulator of NF-κB and cell death, in the immune system. Immunol. Rev. 266, 175–189 (2015).
Takiuchi, T. et al. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19, 254–272 (2014).
Damgaard, R. B. et al. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230.e20 (2016).
Elliott, P. R. & Komander, D. Regulation of Met1-linked polyubiquitin signalling by the deubiquitinase OTULIN. FEBS J. 283, 39–53 (2016).
Heger, K. et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018).
Fujita, H. et al. Mechanism underlying IkB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol. Cell. Biol. 34, 1322–1335 (2014).
Fujita, H. et al. Cooperative domain formation by homologous motifs in HOIL-1L and SHARPIN plays a crucial role in LUBAC stabilization. Cell Rep. 23, 1192–1204 (2018).
Boisson, B. et al. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212, 939–951 (2015).
Yang, Y. et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov. 4, 480–493 (2014).
Elton, L., Carpentier, I., Verhelst, K., Staal, J. & Beyaert, R. The multifaceted role of the E3 ubiquitin ligase HOIL-1: beyond linear ubiquitination. Immunol. Rev. 266, 208–221 (2015).
Brazee, P., Dada, L. A. & Sznajder, J. I. Role of linear ubiquitination in health and disease. Am. J. Respir. Cell Mol. Biol. 54, 761–768 (2016).
Boisson, B. et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186 (2012).
Noad, J. et al. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2, 17063 (2017).
Sakamoto, H. et al. Gliotoxin suppresses NF-κB activation by selectively inhibiting linear ubiquitin chain assembly complex (LUBAC). ACS Chem. Biol. 10, 675–681 (2015).
van Wijk, S. J. L. et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2, 17066 (2017).
MacDuff, D. A. et al. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife https://doi.org/10.7554/eLife.04494.001 (2015).
de Jong, M. F., Liu, Z., Chen, D. & Alto, N. M. Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat. Microbiol. 1, 16084 (2016).
Stieglitz, B., Morris-Davies, A. C., Koliopoulos, M. G., Christodoulou, E. & Rittinger, K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 (2012).
Smit, J. J. et al. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J. Biol. Chem. 288, 31728–31737 (2013).
Tokunaga, F. et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Cohen, P., Kelsall, I. R., Nanda, S. K. & Zhang, J. HOIL-1, an atypical E3 ligase that controls MyD88 signalling by forming ester bonds between ubiquitin and components of the Myddosome. Adv. Biol. Regul. 75, 100666 (2020).
Kelsall, I. R., Zhang, J., Knebel, A., Arthur, J. S. C. & Cohen, P. The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc. Natl Acad. Sci. USA 116, 13293–13298 (2019).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011).
Dove, K. K. & Klevit, R. E. RING-Between-RING E3 ligases: emerging themes amid the variations. J. Mol. Biol. 429, 3363–3375 (2017).
Peltzer, N., Darding, M. & Walczak, H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell. Biol. 26, 445–461 (2016).
Ting, A. T. & Bertrand, M. J. M. More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545 (2016).
Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009).
Peltzer, N. et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 (2014).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Reiter, K. H. & Klevit, R. E. Characterization of RING-Between-RING E3 ubiquitin transfer mechanisms. Methods Mol. Biol. 1844, 3–17 (2018).
Walden, H. & Rittinger, K. RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nat. Struct. Mol. Biol. 25, 440–445 (2018).
Dove, K. K. et al. Structural studies of HHARI/UbcH7~Ub reveal unique E2~Ub conformational restriction by RBR RING1. Structure 25, 890–900.e5 (2017).
Maes, M., Vinken, M. & Jaeschke, H. Experimental models of hepatotoxicity related to acute liver failure. Toxicol. Appl. Pharmacol. 290, 86–97 (2016).
Mignon, A. et al. LPS challenge in d-galactosamine-sensitized mice accounts for caspase-dependent fulminant hepatitis, not for septic shock. Am. J. Respir. Crit. Care Med. 159, 1308–1315 (1999).
Kumari, S. et al. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. eLife https://doi.org/10.7554/eLife.03422.001 (2014).
Stieglitz, B. et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 503, 422–426 (2013).
Dove, K. K. et al. Two functionally distinct E2/E3 pairs coordinate sequential ubiquitination of a common substrate in Caenorhabditis elegans development. Proc. Natl Acad. Sci. USA 114, E6576–E6584 (2017).
Scott, D. C. et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214.e24 (2016).
Dubois, S. M. et al. A catalytic-independent role for the LUBAC in NF-κB activation upon antigen receptor engagement and in lymphoma cells. Blood 123, 2199–2203 (2014).
MacKay, C. et al. E3 ubiquitin ligase HOIP attenuates apoptotic cell death induced by cisplatin. Cancer Res. 74, 2246–2257 (2014).
Niu, J., Shi, Y., Iwai, K. & Wu, Z. H. LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 30, 3741–3753 (2011).
Klein, T. et al. The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat. Commun. 6, 8777 (2015).
Douanne, T., Gavard, J. & Bidere, N. The paracaspase MALT1 cleaves the LUBAC subunit HOIL1 during antigen receptor signaling. J. Cell Sci. 129, 1775–1780 (2016).
Hailfinger, S. et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl Acad. Sci. USA 106, 19946–19951 (2009).
Wang, K. et al. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 5, 67 (2013).
Nilsson, J. et al. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann. Neurol. 74, 914–919 (2013).
Krenn, M. et al. Mutations outside the N-terminal part of RBCK1 may cause polyglucosan body myopathy with immunological dysfunction: expanding the genotype–phenotype spectrum. J. Neurol. 265, 394–401 (2018).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Ohtake, F., Saeki, Y., Ishido, S., Kanno, J. & Tanaka, K. The K48-K63 branched ubiquitin chain regulates NF-κB signaling. Mol. Cell 64, 251–266 (2016).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
van Wijk, S. J., Fiskin, E. & Dikic, I. Selective monitoring of ubiquitin signals with genetically encoded ubiquitin chain-specific sensors. Nat. Protoc. 8, 1449–1458 (2013).
Matsumura, H., Hasuwa, H., Inoue, N., Ikawa, M. & Okabe, M. Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem. Biophys. Res. Commun. 321, 275–279 (2004).
Tamiya, H. et al. IFN-γ or IFN-α ameliorates chronic proliferative dermatitis by inducing expression of linear ubiquitin chain assembly complex. J. Immunol. 192, 3793–3804 (2014).
Acknowledgements
We thank Y. Takeda, T. Jo and Y. Sasaki for insightful discussion, T. Nakagawa and J. Matsuhiro for protein purification, Y. Sugahara and Y. Hayamizu for animal care, and N. Ueno for technical assistance. This study was supported by the following: JSPS KAKENHI grant numbers 24112002, 25253019, JP17H06174 and JP18H05499 (to K.I.); JSPS KAKENHI grant number JP17K08786, a Grant for Joint Research Project of the Institute of Medical Science from the University of Tokyo, and the Hakubi-project grant, Kyoto University (to M.K.); and JSPS KAKENHI grant number JP18H05498 (to F.O.).
Author information
Authors and Affiliations
Contributions
Y.F., H.F. and K.I. conceived and designed the project. Y.F. performed most of the experiments. M.K. and A.N. performed the bacterial infection experiments. F.O., Y.S. and K.T. performed the MS experiments. K.S. performed immunostaining of lungs of mice and generation of M1-TUBE. R.T. provided advice on the project. Y.F. and K.I. wrote the manuscript with contributions from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The highly conserved HOIL-1L RBR ubiquitin ligase negatively regulates LUBAC functions.
a, Conserved residues in the RING1-IBR-RING2 domains of HOIL-1L in the indicated species. Arrow indicates the catalytic cysteine of HOIL-1L. b, Schematic representation of mouse HOIL-1L and its mutants. Cleavage of caspase-3 in LUBAC TKO MEFs stably reconstituted with HOIP and the indicated HOIL-1L protein, stimulated with TNF-α (3 ng ml-1) and CHX (20 μg ml-1), was assessed by immunoblotting. The experiments were repeated twice, independently, with similar results. c, Viability of LUBAC TKO MEFs stably reconstituted with HOIP, SHARPIN, and the indicated HOIL-1L protein, stimulated with TNF-α (10 ng ml-1), was measured using the iCELLigence system. The experiments were repeated three times, independently, with similar results. d, NF-κB activation in HEK293T cells transfected with the indicated expression plasmids and 5× NF-κB luciferase reporters was measured by luciferase assay. Mean is shown; n=3 independent experiments; P-values are from a two-tailed Student’s t-test. e, Quantitative PCR (qPCR) analyses of expression levels of NF-κB target genes in LUBAC TKO MEFs stably reconstituted with HOIP, SHARPIN, and the indicated HOIL-1L protein. Mean ± S.D. is shown; n=5 independent experiments; P-values are from a two-tailed Student’s t-test. f, Phosphorylation and degradation of IκBα in LUBAC TKO MEFs stably reconstituted with HOIP and SHARPIN. The experiments were repeated three times, independently, with similar results. Statistical source data are provided in Source Data Extended Data Fig. 1 and unprocessed immunoblots are provided in Source Data Extended Data Fig. 1.
Extended Data Fig. 2 Loss of HOIL-1L E3 does not overtly affect K48, K63, or other types of chains.
a, Amounts of the indicated types of ubiquitin chains in LUBAC TKO MEFs stably reconstituted with HOIP and SHARPIN, and the indicated HOIL-1L protein. Lysates from TKO MEFs expressing the indicated HOIL-1L proteins were probed with the indicated antibodies. The experiments were repeated three times, independently, with similar results. b, Assessment of ubiquitin linkages in lysates of LUBAC TKO MEFs stably reconstituted with HOIP, SHARPIN, and the indicated HOIL-1L protein using MS analyses. Mean is shown; n=3 independent experiments; P-values were obtained by one-way ANOVA followed by Tukey’s multiple comparison test. Statistical source data are provided in Source Data Extended Data Fig. 2 and unprocessed immunoblots are provided in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 Auto-ubiquitination of HOIL-1L inhibits LUBAC functions.
a, Lysates of LUBAC TKO MEFs stably reconstituted with HOIP C879A, SHARPIN, and the indicated HOIL-1L protein were probed as depicted. Arrow shows the upper band of HOIL-1L. b, In vitro ubiquitination reactions were performed at 37°C for 24 h with His-UBCH7 C/S to generate an oxy-ester bond between ubiquitin and UBCH7, followed by digestion of ubiquitinated UBCH7 with NH2OH or USP2cc at 37°C for 1 h. c,h,l Lysates of indicated MEFs were probed as depicted. d,i Digestion of the upper (modified) HOIL-1L signal with USP2cc or NH2OH at 37°C for 30 min anti-FLAG immunoprecipitates obtained under denaturing conditions from LUBAC TKO MEFs expressing 3×FLAG–HOIL-1L WT (d) or the All-K/R mutant (i). e,j,k, LUBAC TKO MEFs stably reconstituted with HOIP, SHARPIN and 3×FLAG-HOIL WT (e) or the All-K/R mutant (j,k) were subjected to mass spectrometry to assess ubiquitination sites in LUBAC subunits. S107 of hHOIP (e), S915 of mHOIP (j), and T955 of mHOIP (k) were identified as ubiquitination sites. f, Lysates of HEK293T cells transfected with the indicated expression plasmids were probed as indicated. g, NF-kB activation in HEK293T cells transfected with the indicated expression plasmids and 5×NF-kB luciferase reporters was assessed by luciferase assay. Mean is shown; n=3 independent experiments; P-values are from a two-tailed Student’s t-test. The data shown were repeated twice, independently, with similar results. Statistical source data are provided in Source Data Extended Data Fig. 3 and unprocessed immunoblots are provided in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 Loss of HOIL-1L E3 activity increases LUBAC activity in vitro.
a, b, Linear ubiquitin (a) or modification of indicated HOIL-1L proteins (b) in in vitro ubiquitination assays with Petit-SHARPIN and HOIL-1L WT or the indicated mutants, as assessed by immunoblotting. The data shown were repeated three times, independently, with similar results. Unprocessed immunoblots are provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 HOIL-1L ubiquitinates all LUBAC subunits, HOIP conjugates linear ubiquitin onto the ubiquitin, and OTULIN counteracts this effect.
a, Modification of LUBAC subunits in WT or OTULIN KO MEFs treated or not treated with lysates containing MBP-OTULIN at 37°C for 1 h. * shows non-specific bands. The experiments were repeated three times, independently, with similar results. b, Schematic representation of HOIL-1L WT and ΔRING1 (lacking aa 251–341). c, Cleavage of caspase-8 and caspase-3 induced by TNF-a (1 ng ml-1) plus CHX (20 μg ml-1) in WT or OTULIN KO MEFs expressing the indicated HOIL-1L proteins. The experiments were repeated twice, independently, with similar results. Unprocessed immunoblots are provided in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 Loss of HOIL-1L E3 protects against Salmonella typhimurium infection, and infecting S. typhimurium are covered with high levels of linear ubiquitin in MEFs expressing ligase-defective HOIL-1L.
a-d, S. typhimurium infections of LUBAC TKO MEFS stably expressing HOIP, SHARPIN, and 3×FLAG–HOIL-1L WT or C458A. Microscopic images of indicated MEFs were collected 24 hours after infection. Scale bars, 500 μm (a). Confocal micrographs of LUBAC TKO MEFs stably expressing HOIP, SHARPIN, and 3×FLAG–HOIL-1L WT or C458A were infected with S. typhimurium and stained for linear ubiquitin (1E3), FLAG (M2), or Hoechst 33342 at 2 h (b), 4 h (c), and 6 h (d) post-infection. Scale bars, 5 μm (b–d). The data shown were repeated three times, independently, with similar results.
Extended Data Fig. 7 Generation of HOIL-1L ΔRING1 mice as a model for ligase-defective HOIL-1L mice.
a, LUBAC TKO MEFs stably reconstituted with the indicated proteins were probed as shown. b, NF-kB activation in HEK293T cells transfected with the indicated expression plasmids and 5× NF-kB luciferase reporters was assessed by luciferase assay. Mean ± S.D. is shown; n=6 independent experiments; P-values are from one-way ANOVA followed by Tukey’s multiple comparison test. c, Cleavage caspase-3 in LUBAC TKO MEFs stably reconstituted with the indicated proteins and stimulated with TNF-a (2.5 ng ml-1) plus CHX (20 μg ml-1) was assessed by immunoblotting. d, Level of linear ubiquitin in LUBAC TKO MEFs stably reconstituted with the indicated proteins. e, Schematic representation of conditional and deleted loci of HOIL-1L conditional ΔRING1 mice. f, Cell lysates of primary MEFs from WT and HOIL-1L ΔRING1 mice were probed as indicated. g, Indicated MEFs were immunoprecipitated with anti-SHARPIN antibody, and bound and unbound fractions were probed as indicated. h, Lysates of the indicated organs from 8-week-old littermate mice of the indicated genotypes were probed as depicted. i, Splenocytes of 15-week-old littermate mice of the indicated genotypes were examined by flow cytometry. Splenocytes were analyzed for surface expression of CD3e and CD19 (upper panels). B cells were defined as CD19+CD3e- cells and T cells were defined as CD19−CD3e+ cells. T cell subpopulations of splenocytes were analyzed for surface expression of CD4 and CD8 (lower panels). Figures exemplifying the gating strategy for flow cytometry experiments were provided in Supplementary Figure 1c. Experiments were repeated independently with similar results twice (a,c,g,h,i) or at least three times (d,f). Statistical source data are provided in Source Data Extended Data Fig. 7 and unprocessed immunoblots are provided in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Pathophysiological roles of HOIL-1L ΔRING1 in mice.
a, Primary MEFs from WT and HOIL-1L ΔRING1 mice were probed as depicted after stimulation with TNF-a (5 ng ml-1) plus CHX (20 μg ml-1) for the indicated periods. b, Lysates of primary hepatocytes from 12-week-old WT or HOIL-1L ΔRING1 mice were probed as indicated. c, Cleavage of caspase-3 in primary hepatocytes from 12-week-old WT or HOIL-1L ΔRING1 mice after stimulation with TNF-a (10 ng ml-1) plus actinomycin D (Act-D) (100 ng ml-1) was assessed by immunoblotting. d, Macroscopic appearance of livers of the indicated mice at 18 weeks old. Scale bar, 1 cm. e,f, Mice of the indicated genotypes were injected intraperitoneally with LPS/D-GalN or PBS. Seven hours after injection, DNA ladder (e) or cleaved caspase-8 (f) in liver lysates of the indicated mice was probed as depicted. g, Primary keratinocytes from 7-week-old WT or HOIL-1L ΔRING1 mice were probed as indicated. h, Cleavage of caspase-3 in primary keratinocytes from 7-week-old WT or HOIL-1L ΔRING1 mice after stimulation with TNF-a (10 ng ml-1) plus CHX (20 μg ml-1) was assessed by immunoblotting. Experiments were repeated independently with similar results twice (a,c,e-h) or at least three times (b,d). Unprocessed immunoblots are provided in Source Data Extended Data Fig. 8.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Table 1: List of antibodies used; Supplementary Table 2: List of primers using for qRT–PCR gene expression analysis.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots
Source Data Fig. 5
Unprocessed western blots
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Fuseya, Y., Fujita, H., Kim, M. et al. The HOIL-1L ligase modulates immune signalling and cell death via monoubiquitination of LUBAC. Nat Cell Biol 22, 663–673 (2020). https://doi.org/10.1038/s41556-020-0517-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0517-9
This article is cited by
-
HOIL-1L deficiency induces cell cycle alteration which causes immaturity of skeletal muscle and cardiomyocytes
Scientific Reports (2024)
-
Myofiber-type-dependent ‘boulder’ or ‘multitudinous pebble’ formations across distinct amylopectinoses
Acta Neuropathologica (2024)
-
OTULIN Can Improve Spinal Cord Injury by the NF-κB and Wnt/β-Catenin Signaling Pathways
Molecular Neurobiology (2024)
-
Biallelic human SHARPIN loss of function induces autoinflammation and immunodeficiency
Nature Immunology (2024)
-
Exploiting E3 ubiquitin ligases to reeducate the tumor microenvironment for cancer therapy
Experimental Hematology & Oncology (2023)