Abstract
The crosstalk between deregulated hepatocyte metabolism and cells within the tumour microenvironment, as well as the consequent effects on liver tumorigenesis, are not completely understood. We show here that hepatocyte-specific loss of the gluconeogenic enzyme fructose 1,6-bisphosphatase 1 (FBP1) disrupts liver metabolic homeostasis and promotes tumour progression. FBP1 is universally silenced in both human and murine liver tumours. Hepatocyte-specific Fbp1 deletion results in steatosis, concomitant with activation and senescence of hepatic stellate cells (HSCs), exhibiting a senescence-associated secretory phenotype. Depleting senescent HSCs by ‘senolytic’ treatment with dasatinib/quercetin or ABT-263 inhibits tumour progression. We further demonstrate that FBP1-deficient hepatocytes promote HSC activation by releasing HMGB1; blocking its release with the small molecule inflachromene limits FBP1-dependent HSC activation, the subsequent development of the senescence-associated secretory phenotype and tumour progression. Collectively, these findings provide genetic evidence for FBP1 as a metabolic tumour suppressor in liver cancer and establish a critical crosstalk between hepatocyte metabolism and HSC senescence that promotes tumour growth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The human liver cancer data were derived from the TCGA Research Network (https://cancergenome.nih.gov/cancersselected/LiverHepatocellularCarcinoma). Two public mouse NAFLD-HCC datasets (GSE67680, ref. 19, and GSE99010, ref. 20) are available at https://www.ncbi.nlm.nih.gov/geo/. The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus with the accession number GSE135616. The mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD017831. The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary information files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The software and algorithms for data analyses used in this study are all well established from previous work and are referenced throughout the manuscript. No custom code was used in this study.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017).
The Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341 (2017).
Greten, T. F., Wang, X. W. & Korangy, F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut 64, 842–848 (2015).
Ringelhan, M., Pfister, D., O’Connor, T., Pikarsky, E. & Heikenwalder, M. The immunology of hepatocellular carcinoma. Nat. Immunol. 19, 222–232 (2018).
Friedman, S. L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 88, 125–172 (2008).
Affo, S., Yu, L. X. & Schwabe, R. F. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu. Rev. Pathol. 12, 153–186 (2017).
Gomes, A. L. et al. Metabolic inflammation-associated Il-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell 30, 161–175 (2016).
Ma, C. et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature 531, 253–257 (2016).
Shalapour, S. et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 551, 340–345 (2017).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
Arab, J. P., Arrese, M. & Trauner, M. Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annu. Rev. Pathol. 13, 321–350 (2018).
Michelotti, G. A., Machado, M. V. & Diehl, A. M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 10, 656–665 (2013).
Dong, C. et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23, 316–331 (2013).
Hirata, H. et al. Decreased expression of fructose-1,6-bisphosphatase associates with glucose metabolism and tumor progression in hepatocellular carcinoma. Cancer Res. 76, 3265–3276 (2016).
Li, B. et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 513, 251–255 (2014).
Gebhardt, R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol. Ther. 53, 275–354 (1992).
Katz, S. F. et al. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology 142, 1229–1239 (2012).
Tschaharganeh, D. F. et al. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell 158, 579–592 (2014).
Asgharpour, A. et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol. 65, 579–588 (2016).
Tsuchida, T. et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 69, 385–395 (2018).
Arruda, A. P. et al. Chronic enrichment of hepatic endoplasmic reticulum–mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 20, 1427–1435 (2014).
Febbraio, M. A. et al. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 29, 18–26 (2019).
Nakagawa, H. et al. ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 26, 331–343 (2014).
Bakiri, L. & Wagner, E. F. Mouse models for liver cancer. Mol. Oncol. 7, 206–223 (2013).
He, G. et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell 155, 384–396 (2013).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Schnabl, B., Purbeck, C. A., Choi, Y. H., Hagedorn, C. H. & Brenner, D. Replicative senescence of activated human hepatic stellate cells is accompanied by a pronounced inflammatory but less fibrogenic phenotype. Hepatology 37, 653–664 (2003).
Henderson, N. C. et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 (2013).
Baar, M. P. et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147 (2017).
Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010).
Zhu, Y. et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015).
Farr, J. N. et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 23, 1072–1079 (2017).
Ogrodnik, M. et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 8, 15691 (2017).
Xu, M. et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 24, 1246–1256 (2018).
Zhu, Y. et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 15, 428–435 (2016).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Guieze, R. et al. Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell 36, 369–384 (2019).
Hernandez, C. et al. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J. Clin. Invest. 128, 2436–2451 (2018).
Khambu, B. et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J. Clin. Invest. 128, 2419–2435 (2018).
Ge, X. et al. High mobility group box-1 drives fibrosis progression signaling via the receptor for advanced glycation end products in mice. Hepatology 68, 2380–2404 (2018).
Chen, R. et al. Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol. Med. 19, 357–366 (2013).
Kim, K. H., Chen, C. C., Monzon, R. I. & Lau, L. F. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol. Cell. Biol. 33, 2078–2090 (2013).
Seki, E. et al. CCR2 promotes hepatic fibrosis in mice. Hepatology 50, 185–197 (2009).
Lee, S. et al. A small molecule binding HMGB1 and HMGB2 inhibits microglia-mediated neuroinflammation. Nat. Chem. Biol. 10, 1055–1060 (2014).
Huangyang, P. et al. Fructose-1,6-bisphosphatase 2 inhibits sarcoma progression by restraining mitochondrial biogenesis. Cell Metab. 31, 174–188 (2019).
Lee, S. & Schmitt, C. A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 21, 94–101 (2019).
Eggert, T. et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell 30, 533–547 (2016).
Kang, T. W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
Lujambio, A. et al. Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460 (2013).
Liu, F. et al. Ceramide activates lysosomal cathepsin B and cathepsin D to attenuate autophagy and induces ER stress to suppress myeloid-derived suppressor cells. Oncotarget 7, 83907–83925 (2016).
Tian, Z., Chen, Y. & Gao, B. Natural killer cells in liver disease. Hepatology 57, 1654–1662 (2013).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Baker, D. J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Kirkland, J. L. & Tchkonia, T. Cellular senescence: a translational perspective. EBioMedicine 21, 21–28 (2017).
Kirkland, J. L., Tchkonia, T., Zhu, Y., Niedernhofer, L. J. & Robbins, P. D. The clinical potential of senolytic drugs. J. Am. Geriatr. Soc. 65, 2297–2301 (2017).
Childs, B. G. et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 (2017).
Dorr, J. R. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013).
Toso, A. et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 9, 75–89 (2014).
Demaria, M. et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017).
Wang, C. et al. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature 574, 268–272 (2019).
Mu, X. et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J. Clin. Invest. 125, 3891–3903 (2015).
Mederacke, I., Dapito, D. H., Affo, S., Uchinami, H. & Schwabe, R. F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Riscal, R. et al. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol. Cell 62, 890–902 (2016).
Possemato, R. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011).
Acknowledgements
We thank the members of the Simon laboratory for their helpful discussions and insights on the manuscript. We also thank S. Berger for critical reading of the manuscript and K. E. Wellen for providing the D37 cells. H.-Y. Tang assisted in hepatocyte secretome profiling and data analysis. C. Mesaros and L. Weng assisted in lipid profiling and data analysis. We are grateful to J. Tobias for help with processing the human TCGA and mouse RNA-seq data, and A. Durham for histopathologic examination. We also thank W. Quinn III, A. J. Merrell, H. Xie and H. Weinstein for their technical assistance. This work was supported by the National Key Research and Development Program (grant no. 2016YFA0502600) of China (to B.L.) and National Cancer Institute (NCI) grant nos P01CA104838, R35CA197602 and P30CA016520 (to M.C.S.).
Author information
Authors and Affiliations
Contributions
F.L., B.L. and M.C.S. conceived the project and designed the research studies. F.L. performed most of the experiments described. M.B. and K.G. provided help with animal husbandry and technical assistance in the mouse experiments. B.L. performed the Fbp1 gene targeting and generated the chimaeric mice. P.H., R.R., J.G. and I.A.B. assisted with the in vitro assays. K.E.L. and N.L. provided technical assistance for the flow cytometry and data analysis. P.H., P.L. and B.K. provided conceptual advice and helpful discussion. F.L. analysed data. F.L., B.K., B.L. and M.C.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
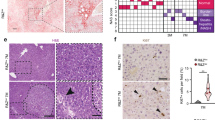
Extended Data Fig. 1 Decreased FBP1 expression in human and murine liver tumours.
a, Metabolic gene set analysis of TCGA HCC RNA-sequencing data. A total of 374 HCC tumours and 50 adjacent normal tissues were included, and 2752 genes encoding all known human metabolic enzymes and transporters were classified according to the Kyoto Encyclopedia of Genes and Genomes (KEGG). Generated metabolic gene sets were ranked based on their median fold expression changes in HCC tumours vs normal tissue, and plotted as median ± median absolute deviation. b, Representative FBP1 IHC staining on human liver tissue array with adjacent normal, grade 2 and 3 HCC tissues. Scale bar: 100 µm. c, Statistical analysis of FBP1 IHC staining in (b). n=20 for normal, n=30 for grade 2, n=30 for grade 3 samples. In each box plot, the top-most line is the maximum, the top of the box is the third quartile, the centre line is the median, the bottom of the box is the first quartile and the bottom-most line is the minimum. d, Representative H&E staining in liver sections from 24-week control (Ctrl) and DEN-treated (DEN) C57BL/6 mice (n = 3 independent experiments with similar results). T, tumour. Scale bar: 100 µm. e, Serum alanine transaminase (ALT) activity from 24-week Ctrl and DEN mice. n = 4 for Ctrl, n = 5 for DEN. Graph in e show mean ± SEM, and P value was calculated using a two-tailed t-test. Numerical source data are provided in Source Data Extended Data Fig. 1.
Extended Data Fig. 2 Hepatic FBP1 loss disrupts liver metabolism in mice.
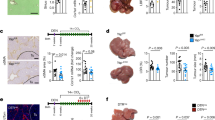
a, Scheme for generating Fbp1fl/fl mice by homologous recombination. b, Scheme for hepatocyte-specific Fbp1 deletion in Fbp1fl/fl mice. c, qRT-PCR analysis of gluconeogenic gene expression in 24-week GFP (n = 4) and Cre (n=4) livers. d, Immunoblotting analysis of 24-week GFP and Cre livers (n = 3 independent experiments). GAPDH was used as loading control. e, H&E staining of 24-week GFP and Cre kidney sections (n = 3 independent experiments). Scale bar: 100 µm. f, Immunoblotting analysis of 24-week GFP (n=2) and Cre (n=2) kidneys. GAPDH was used as loading control. g, qRT-PCR analysis of gluconeogenic gene expression in 24-week GFP (n=4) and Cre (n=4) kidneys. h, Serum free fatty acid (FFA) and β-hydroxybutyrate (BHBA) levels of fasted (16-h) and refed (4-h) GFP (n=7) and Cre (n=8) mice (24-week). i, Liver gross appearance of 16-h fasted animals (24-week) (n = 3 independent experiments). j,k, qRT-PCR analysis of lipid metabolism (j) and unfolded protein response (UPR) (k) gene signatures in 16-h fasted GFP (n = 5) and Cre (n=5) livers (24-week). In each box plot, the top-most line is the maximum, the top of the box is the third quartile, the centre line is the median, the bottom of the box is the first quartile and the bottom-most line is the minimum. l, Growth rates of GFP and Cre mice. GFP: n=5 for female or male mice, Cre: n=5 for female, n = 8 for male mice. m,n, Quantification of triglyceride (TG) (m) and Oil Red O staining (% area) (n) in 24-week GFP (n = 6) and Cre (n = 5) mouse livers. Graphs in c, g, h, l-n show mean ± SEM. All P values were calculated using a two-tailed t-test. Scanned images of unprocessed blots in c and e are shown in Source Data Extended Data Fig. 2. Numerical source data are provided in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 Hepatic FBP1 loss promotes tumour progression in p53fl/fl and NAFLD models.
a, Scheme for Fbp1 deletion in DEN-induced liver cancer model. b, Gross liver appeara = nce and tumour multiplicity in 80-week p53 and p53/Fbp1 mice treated with AAV8-TBG-Cre. Yellow arrows indicate liver tumours. Scale bar: 1 cm. c, Quantification of surface tumour numbers in p53 and p53/Fbp1 animals in (b). n = 7 mice for p53 or p53/Fbp1 cohorts. 4 of 7 p53 mice and 7 of 7 p53/Fbp1 mice had surface tumours. d, Representative H&E staining and α-SMA IHC staining of liver sections from 80-wk p53 and p53/Fbp1 mice (n = 3 independent experiments). Scale bar: 100 µm. e, Gross liver appearances of 32-wk GFP and Cre mice with diet- and CCl4-induced NAFLD (see Materials and Methods for details). Scale bar: 1 cm. (f-i) Quantification of surface tumour number (f), liver-to-body weight ratio (g), liver weight (h) and body weight (i) in 32-wk GFP (n = 5) and Cre (n = 5) mice with NAFLD. j,k, qRT-PCR analysis of lipogenic (j) and fibrotic (k) gene expression from 32-wk GFP (n = 5) and Cre (n = 5) mouse livers with NAFLD. l, Representative H&E staining, Sirius Red staining of 32-wk GFP and Cre NAFLD mouse liver sections (n = 3 independent experiments). Scale bar: 100 µm. m, Quantification of Sirius Red staining in (l). n = 5 mice for each group. All graphs represent the mean ± SEM. In each box plot of j, k and m, the top-most line is the maximum, the top of the box is the third quartile, the centre line is the median, the bottom of the box is the first quartile and the bottom-most line is the minimum. Graphs in c, f-I show mean ± SEM. All P values were calculated using a two-tailed t-test. Numerical source data are provided in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 Impact of hepatic FBP1 loss on tumour microenvironment in DEN mice.
a, Representative CYP2E1 IHC staining in liver sections from 24-week DEN/GFP and DEN/Cre mice (n = 3 independent experiments). Scale bar: 100 µm. b, Flow cytometry quantification of T cell subpopulations in 24-week DEN/GFP (n = 4) and DEN/Cre (n = 5) livers. c, Flow cytometry quantification of B cells and B cell subpopulations in 24-week DEN/GFP (n = 4) and DEN/Cre (n = 5) livers. d, Flow cytometry quantification of total macrophages and CD11b+ or CD206+ subsets in 24-week DEN/GFP (n = 4) and DEN/Cre (n = 5) livers. e,f, Representative flow cytometry plots (e) and quantification (f) (% CD45+ cells) of NK cells (CD3−NKp46+) in 24-week DEN/GFP (n = 4) and DEN/Cre (n = 5) livers. g,h, Representative NKp46 IHC staining (g) and quantification (h) in 24-wk DEN/GFP (n = 4) and DEN/Cre (n = 5) liver sections. Scale bar: 100 µm. i,j, Representative flow cytometry plots (i) and quantification (j) (% CD45+ cells) of MDSC cells (CD11b+Ly6C+) in 24-week DEN/GFP (n = 4) and DEN/Cre (n = 5) livers. k, A heatmap showing relative abundance of individual ceramide species in 24-wk DEN/GFP (N = 7) and DEN/Cre (n = 9) mouse livers by lipidomic profiling. Graphs in b-d, f, h and j show mean ± SEM, and P values were calculated using a two-tailed t-test. Numerical source data are provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 Hepatic FBP1 loss leads to HSC activation and senescence.
a, Representative SA-β-Gal staining, α-SMA and IL6 IHC staining of serial cryosections from 36-week mouse livers (n=3 independent experiments). Scale bar: 100 µm. b, Representative Sirius Red staining and quantification, Ki67/α-SMA IF staining and quantification of 24-week non-DEN liver sections. For Sirius Red staining quantification, n = 20 fields of view (FOV, 200x) from 6 mice for GFP, n = 18 fields of view (FOV, 200x) from 6 mice for Cre. For Ki67/α-SMA IF staining quantification, n = 6 mice for each group. Scale bar: 100 µm. c, SA-β-Gal staining and quantification (% of cells) in 24-week liver sections from non-DEN mice. Black arrows indicate SA-β-Gal staining. n = 6 mice for each group. Scale bar: 100 µm. d, α-SMA and γ-H2AX IHC staining of 24-week non-DEN Cre liver sections. Scale bar: 100 µm. e, Quantification of α-SMA/γ-H2AX IHC staining and SASP component IF staining of 24-week non-DEN Cre (n = 6) liver sections. Graphs in b, c and e show mean ± SEM, and P values were calculated using a two-tailed t-test. Numerical source data are provided in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 In vitro Characterization of senolytic effects of D+Q and ABT-263.
a, Growth curves of GRO and SEN human HSCs under 3% O2 (to prevent senescence due to oxidative damage) in regular medium (n = 3 independent experiments). b, Growth curves of mouse D37 cells under 3% O2 in conditioned medium from Vehicle (Veh) or etoposide (Etp)-treated mouse HSCs (n = 3 independent experiments). c, Viability or apoptosis (% Annexin V+) quantification of GRO or SEN human HSCs after D+Q treatment at indicated concentrations/combinations (n = 3 or independent experiments). d, Viability or apoptosis (% Annexin V+) quantification of GRO or SEN human HSCs after treatment with ABT-263 at indicated concentrations (n = 3 independent experiments). e, Apoptosis (% Annexin V+) quantification of GRO or SEN human HSCs after treatment with indicated AZD5991 (50 nM) and ABT-263 (10 μm) combinations (n = 3 independent experiments). f, Viability or apoptosis (% Annexin V+) quantification of mouse D37 cells after D+Q treatment at indicated concentrations/combinations (n = 3 independent experiments). g, Mouse D37 cell viability or apoptosis (% Annexin V+) quantification after treatment with indicated ABT-263 concentrations (n = 3 independent experiments). All graphs show mean ± SEM, and P values were calculated using a two-tailed t-test. Numerical source data are provided in Source Data Extended Data Fig. 6.
Extended Data Fig. 7 Characterizing the senolytic effects of D+Q and ABT-263 in vivo.
a, Scheme of early stage Veh and D+Q treatment of DEN/GFP or DEN/Cre mice. b,c, Liver-to-Body Weight (LW/BW) ratio (b) and body weight (c) quantifications of 24-week Veh (n = 5) and D+Q (n = 6) DEN/Cre mice. d, Quantification of TG levels from 24-week Veh (n = 5) and D+Q (n = 6) DEN/Cre mouse livers. e,f, Representative Sirius Red staining (e) and quantification (f) of 24-week Veh (n = 25) and D+Q (n = 20) DEN/Cre mouse liver sections. FOV: 200x fields of view. Scale bar: 100 µm. g, TUNEL staining of 24-week Veh and D+Q DEN/Cre mouse liver sections (n = 3 independent experiments). Scale bar: 100 µm. h, Scheme of late stage Veh and D+Q treatment of DEN/GFP or DEN/Cre mice. i, Representative SA-β-Gal staining (n = 3 independent experiments), and BrdU/α-SMA IF staining of 36-week Veh and D+Q DEN/Cre mouse liver sections. Scale bar: 100 µm. j, Quantification of BrdU and α-SMA IF staining of 36-week Veh (n = 5) and D+Q (n = 6) DEN/Cre mouse liver sections. k, Surface tumour number and size distributions of 24 wk or 36 wk DEN/GFP mice treated with Veh or D+Q. n = 5 mice for each cohort at each time point. l, Scheme of Veh and ABT-263 treatment of DEN/GFP or DEN/Cre mice. m, Representative TUNEL staining of 36-week Veh and ABT-263-treated DEN/Cre mouse liver sections (n = 3 independent experiments). Scale bar: 100 µm. n, SA-β-Gal staining of 36-week Veh and ABT-263-treated DEN/Cre mouse liver sections (n = 3 independent experiments). Scale bar: 100 µm. o, Surface tumour number and size distributions of DEN/GFP mice treated with Veh (n=5) or ABT-263 (n = 5). Graphs in b–d, f, j, k and o show mean ± SEM, and P values were calculated using a two-tailed t-test. Numerical source data are provided in Scanned images of unprocessed blots in Source Data Extended Data Fig. 7.
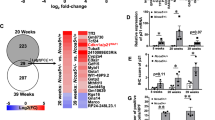
Extended Data Fig. 8 Identification of HMGB1 as a potential mediator between FBP1-deficient hepatocytes and HSCs.
a, Unsupervised hierarchical clustering of normalized protein abundance in CM of 24-week Non-DEN GFP (n = 5) and Cre (n = 5) hepatocytes. b, An Egyptian Pie Chart of 459 proteins with > = 1.5-fold change (adjusted p<0.05) of abundance in CM between Non-DEN GFP (n = 5) and Cre (n = 5) groups. c, ELISA-based quantification of HMGB1 levels in CM of 24-week Non-DEN GFP (n = 4) or Cre (n = 4) hepatocytes. d, Immunoblotting analysis of HMGB1 in the nuclear (Nuc) and cytosolic (Cyto) fractions or total lysates from 24-week non-DEN GFP (n = 2) and Cre (n = 2) mouse livers. H3 and HSP90 were used as loading control for nuclear and cytosolic fractions, respectively. GAPDH was used as loading control for whole tissue lysates. e, qRT-PCR analysis of UPR gene expression in mouse primary hepatocytes after tunicamycin (TUN) treatment (n = 3 independent experiments). f, ELISA-based quantification of HMGB1 levels in primary hepatocyte culture medium of Ctrl and TUN groups (n = 3 independent experiments). g,h, qRT-PCR analysis of SASP (g) or fibrotic (h) gene expression in human HSCs after 1 nM HMGB1 treatment for 15 h (n = 3 independent experiments). Graphs in c, e, f-h show represent the mean ± SEM, and P values were calculated using a two-tailed t-test. Scanned images of unprocessed blots in d are shown in Source Data Extended Data Fig. 8. Numerical source data are provided in Source Data Extended Data Fig. 8.
Extended Data Fig. 9 Characterization of in vivo and in vitro ICM treatment.
a, Scheme for Veh and ICM treatment of DEN/GFP and DEN/Cre mice. b, Gross liver appearances and tumour multiplicity (indicated by yellow arrows) in Veh and ICM DEN/Cre mice. Scale bar: 1 cm. c, H&E staining of Veh (n = 6) and ICM (n = 7) DEN/Cre mouse liver sections. Scale bar: 100 µm. d, Quantification of TG levels in Veh (n = 6) and ICM (n = 7) DEN/Cre mouse livers. e, Immunoblotting analysis of HMGB1 in nuclear (Nuc) and Cytosolic (Cyto) fractions of Veh (n = 2) and ICM (n = 2) DEN/Cre livers. H3 and HSP90 were used as loading control for nuclear and cytosolic fractions, respectively. f, SA-β-Gal staining of Veh (n = 6) and ICM (n = 7) DEN/Cre mouse liver sections. Scale bar: 100 µm. g, TUNEL staining and quantification of Veh (n = 6) and ICM (n = 6) DEN/Cre mouse liver sections. Scale bar: 100 µm. h, Cell viability assays of GRO human HSCs after ICM treatment (n = 3 independent experiments). I,j, Cell viability assays in SEN human HSCs (i) or mouse D37 cells (j) after ICM (10 μm) treatment (n = 3 independent experiments). k, qRT-PCR analysis of SASP gene expression in human HSCs after 10 µm ICM treatment for 24 h (n = 3 independent experiments). l, Representative Sirius Red staining and quantification (% area) of Veh (n = 25 FOV) and ICM (n = 21) DEN/Cre mouse liver sections. FOV: fields of view. Scale bar: 100 µm. m, Quantification of surface tumour number and size distributions from DEN/GFP mice treated with Veh (n = 5) or ICM (n = 5). Graphs in d, g–m show mean ± SEM, and P values were calculated using a two-tailed t-test. Scanned images of unprocessed blots in e are shown in Source Data Extended Data Fig. 9. Numerical source data are provided in Source Data Extended Data Fig. 9.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 2
Unprocessed gels
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 8
Unprocessed gels
Source Data Extended Data Fig. 9
Statistical source data
Source Data Extended Data Fig. 9
Unprocessed gels
Rights and permissions
About this article
Cite this article
Li, F., Huangyang, P., Burrows, M. et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat Cell Biol 22, 728–739 (2020). https://doi.org/10.1038/s41556-020-0511-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0511-2
This article is cited by
-
Fructose-1,6-bisphosphatase 1 dephosphorylates and inhibits TERT for tumor suppression
Nature Chemical Biology (2024)
-
Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer
Nature Cancer (2024)
-
Cell senescence in liver diseases: pathological mechanism and theranostic opportunity
Nature Reviews Gastroenterology & Hepatology (2024)
-
HMGB1 mediates synaptic loss and cognitive impairment in an animal model of sepsis-associated encephalopathy
Journal of Neuroinflammation (2023)
-
The multifunctional protein HMGB1: 50 years of discovery
Nature Reviews Immunology (2023)