Abstract
Interstitial deletion of the long arm of chromosome 5 (del(5q)) is the most common structural genomic variant in myelodysplastic syndromes (MDS)1. Lenalidomide (LEN) is the treatment of choice for patients with del(5q) MDS, but half of the responding patients become resistant2 within 2 years. TP53 mutations are detected in ~20% of LEN-resistant patients3. Here we show that patients who become resistant to LEN harbour recurrent variants of TP53 or RUNX1. LEN upregulated RUNX1 protein and function in a CRBN- and TP53-dependent manner in del(5q) cells, and mutation or downregulation of RUNX1 rendered cells resistant to LEN. LEN induced megakaryocytic differentiation of del(5q) cells followed by cell death that was dependent on calpain activation and CSNK1A1 degradation4,5. We also identified GATA2 as a LEN-responsive gene that is required for LEN-induced megakaryocyte differentiation. Megakaryocytic gene-promoter analyses suggested that LEN-induced degradation of IKZF1 enables a RUNX1–GATA2 complex to drive megakaryocytic differentiation. Overexpression of GATA2 restored LEN sensitivity in the context of RUNX1 or TP53 mutations by enhancing LEN-induced megakaryocytic differentiation. Screening for mutations that block LEN-induced megakaryocytic differentiation should identify patients who are resistant to LEN.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Microarray data were deposited into the GEO (GSE126265). WGS and RNA-seq data were deposited into the European Genome-phenome Archive (EGAS00001004113). ChIP–seq peak data for RUNX1 and GATA2 was downloaded from BloodChIP35 (accessed April 2018). IKZF1 ChIP–seq peaks are available at the GEO (GSE91730). Source data for Figs. 1–5 and Extended Data Figs. 2–8 and 10 are available with the paper. All other data supporting the findings of this study are available from the corresponding author on request.
References
Bejar, R., Levine, R. & Ebert, B. L. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J. Clin. Oncol. 29, 504–515 (2011).
Gohring, G. et al. Patients with del(5q) MDS who fail to achieve sustained erythroid or cytogenetic remission after treatment with lenalidomide have an increased risk for clonal evolution and AML progression. Ann. Hematol. 89, 365–374 (2010).
Talati, C., Sallman, D. & List, A. F. SOHO state of the art and next questions: management of myelodysplastic syndromes with deletion 5q. Clin. Lymphoma Myeloma Leuk. 18, 629–635 (2018).
Kronke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 (2015).
Fang, J. et al. A calcium- and calpain-dependent pathway determines the response to lenalidomide in myelodysplastic syndromes. Nat. Med. 22, 727–734 (2016).
Jadersten, M. et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J. Clin. Oncol. 29, 1971–1979 (2011).
Scharenberg, C. et al. Progression in patients with low- and intermediate-1-risk del(5q) myelodysplastic syndromes is predicted by a limited subset of mutations. Haematologica 102, 498–508 (2017).
Cunningham, L. et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFβ interaction. Proc. Natl Acad. Sci. USA 109, 14592–14597 (2012).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010).
Zhu, Y. X. et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood 118, 4771–4779 (2011).
Matheny, C. J. et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 26, 1163–1175 (2007).
Croce, K. et al. Inhibition of calpain blocks platelet secretion, aggregation, and spreading. J. Biol. Chem. 274, 36321–36327 (1999).
Sekeres, M. A. et al. Relationship of treatment-related cytopenias and response to lenalidomide in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 26, 5943–5949 (2008).
Yoshida, T., Ng, S. Y., Zuniga-Pflucker, J. C. & Georgopoulos, K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7, 382–391 (2006).
Malinge, S. et al. Ikaros inhibits megakaryopoiesis through functional interaction with GATA-1 and NOTCH signaling. Blood 121, 2440–2451 (2013).
von Palffy, S. et al. Dominant-negative IKAROS enhances IL-3-stimulated signaling in wild-type but not BCR-ABL1+ mouse BA/F3 cells. Exp. Hematol. 43, 514–523 (2015).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Tijssen, M. R. et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 20, 597–609 (2011).
Fink, E. C. et al. Crbn I391V is sufficient to confer in vivo sensitivity to thalidomide and its derivatives in mice. Blood 132, 1535–1544 (2018).
Boultwood, J. et al. Gene expression profiling of CD34+ cells in patients with the 5q− syndrome. Br. J. Haematol. 139, 578–589 (2007).
Graubert, T. A. et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS ONE 4, e4583 (2009).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Cingolani, P. et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 3, 35 (2012).
He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016).
Cancer Genome Atlas Research NetworkGenomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Griffith, M. et al. Alternative expression analysis by RNA sequencing. Nat. Methods 7, 843–847 (2010).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Patro, R., Mount, S. M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol. 32, 462–464 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Chacon, D., Beck, D., Perera, D., Wong, J. W. & Pimanda, J. E. BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res. 42, D172–D177 (2014).
Nguyen, T. T. et al. Revealing a human p53 universe. Nucleic Acids Res. 46, 8153–8167 (2018).
Acknowledgements
We thank I. Martinez-Gonzalez for her comments; C. Eaves for providing the MND-IKZF1 and MND-IKZF1-DN plasmids; K. Tohyama for providing the MDS-L cell line; and S. Sung for technical assistance with preparing patient samples for WGS. This research was supported by grants from the Leukemia and Lymphoma Society of Canada, a Terry Fox Research Institute New Frontiers Program Project grant, the Canadian Institute of Health Research, Genome BC, BC Cancer Foundation through the Leukemia and Myeloma Program and Associazione Italiana per la Ricerca sul 248 Cancro (AIRC, Milan, Italy) Project Codes 20125 and 21267.
Author information
Authors and Affiliations
Contributions
A.Karsan conceived and initiated the study and obtained funding. S.M.-H. and A.Karsan designed the project, interpreted data and wrote the paper. S.M.-H., Y.D., J.J., A.M., N.G. and M.J. performed experiments. S.M.-H. analysed data and generated figures. T.R.D., J.P. and J.L. performed bioinformatics analyses. A.Kulasekararaj, A.F.L., M.J., L.M., A.F.L., E.H.-L. and U.P. provided patient samples and edited the paper. M.F. and P.U. provided technical support for animal experiments.
Corresponding author
Ethics declarations
Competing interests
U.P. has received honoraria and research support from Celgene. A.Kulasekararaj received honoraria from Celgene.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
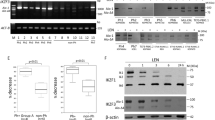
Extended Data Fig. 1 Whole genome and expression analysis in del(5q) MDS at relapse.
a, Copy number analysis of WGS data showing that deletion of the 5q CDR is conserved at relapse in our cohort of del(5q) MDS patients. The x-axis represents the length of chromosome 5 and the y-axis shows the copy number ratio for each chromosomal location. Numbers in red represent mean log2 ratio of chromosome 5q copy number relative to normal. b, Variant allele frequency (VAF) of protein-coding genes in the FoundationOne heme panel at relapse for each patient. c, Relative mRNA expression change of TP53 in this cohort of del(5q) MDS patient marrows at relapse assessed by RT-qPCR. Data is shown as log2fold expression at relapse relative to diagnosis. d,e, Copy number analysis inferred from WGS data for chromosome 17p (d) and chromosome 21 (e). The x-axis represents the length of chromosome and the y-axis shows the copy number ratio for each chromosomal location. RUNX1 and TP53 chromosomal location is marked. The numbers represent mean log ratio of the corresponding gene copy number variation relative to normal.
Extended Data Fig. 2 Lenalidomide activates RUNX1 in a CRBN-dependent manner to eliminate del(5q) MDS cells.
a, b, Growth curves for the del(5q) cell lines MDS-L (a) and KG-1a (n = 3 independent experiments) (b). Graphs represent viable cell counts during the length of treatment with LEN (n = 3 independent experiments). c, Percentage of AnnexinV+ cells after treatment of del(5q) cells with LEN (1 µM) for 7 days (n = 3 independent experiments). d, Colony counts from del(5q) cells plated in methylcellulose supporting myeloerythroid maturation (H4434, STEMCELL Technologies) in the presence of LEN (1 µM) or DMSO control. Total number of colonies were assessed between 7-10 days after plating (n = 3 independent experiments). e, Western blot showing Cereblon expression in control MDS-L and a CRBN-KO clone. IKZF1 and CSNK1A1 protein degradation are shown as a control for LEN binding to Cereblon. GAPDH is shown as loading control. (n = 3 independent experiments). f, Colony counts from control or CRBN-KO clones in the presence of LEN (1 µM) or DMSO control (n = 4 independent experiments). g, CSF1R mRNA expression in control and CRBN-KO cells after treatment with LEN (1 µM) for 24h. (n = 4 independent experiments). h, Western blot showing RUNX1 expression in MDS-L cells transduced with the EGFP empty vector control or FLAG-RUNX1 construct. CBF-b protein expression is shown as loading control. (n = 3 independent experiments). i, Percentage of AnnexinV+ cells after treatment of RUNX1-WT or EGFP overexpressing MDS-L cells five days post-transduction (n = 3 independent experiments). j, Colony counts from RUNX1-WT or EGFP-expressing MDS-L cells in methylcellulose (n = 3 independent experiments). Statistical analysis was performed using two sided Student’s t-test (c,d,I,j) or ANOVA with multiple correction adjustment (a,b,f,g). Data is presented as mean ± s.d. Source data are available online.
Extended Data Fig. 3 Characterization of RUNX1-KO and TP53-KO clones.
a, Sanger sequencing chromatograms showing insertion/deletion generated by CRISPR/Cas9 system in the MDS-L derived clones at the indicated gene. b, Digital droplet PCR values showing CSNK1A1 gene copy number in the clones used in this study. (n = 1 experiment). c, CSF1R mRNA expression in control and RUNX1-KO cells after treatment with LEN (1 µM) for 24h (n = 4 independent experiments). d, Western blot showing p53 protein expression in control and TP53-KO clones. (n = 3 independent experiments) e, Percentage of AnnexinV+ cells after treatment of control or TP53-KO clones after treatment with LEN (1 µM) or DMSO as control for 4 days (n = 3 independent experiments). f, Colony counts from control or TP53-KO clones in the presence of LEN (1 µM) or DMSO control (n = 4 independent experiments). Data from each independent clone is shown in different shades. Statistical analysis was performed using ANOVA with multiple correction adjustment. Data is presented as mean ± s.d. Source data are available online.
Extended Data Fig. 4 RUNX1 downregulation results in resistance to LEN in del(5q) cells.
a,b, Western blot showing downregulation of RUNX1 protein expression in shRNA-expressing cells in KG-1a (a) or MDS-L (b) cells transduced with a lentivirus expressing either shRNA targeting RUNX1 or empty vector control. (n = 3 independent experiments). c,d, Methylcellulose colony counts from KG-1a (c) or MDS-L (d) cells transduced with a lentivirus expressing either shRNA targeting RUNX1 or empty vector control in the presence of LEN (1 µM) or DMSO (n = 3 independent experiments). e, Western blot showing LEN-dependent upregulation of RUNX1 protein levels compared to enforced expression of RUNX1 WT or mutant protein in the RUNX1-KO clone. GAPDH is shown as loading control. f, Quantification of RUNX1 protein levels normalized to GAPDH expression in each lane of (e) was performed using ImageJ (n = 2 independent experiments). Statistical analysis was performed using ANOVA with multiple correction adjustment. Data is presented as mean ± s.d. Source data are available online.
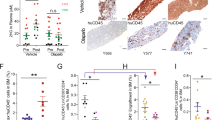
Extended Data Fig. 5 Analysis of MDS-L differentiation in the bone marrow of LEN-treated mice.
a, Differentiation analysis of bone marrow MDS-L engrafted in NRG-3GS mice and treated with LEN in vivo. No significant differentiation is observed in LEN-treated animals compared to DMSO treated for any of the markers shown (n = 3 biological independent samples). b, Representative flow panel showing apoptosis (Annexin V+ cells) occurring mainly in the CD34− compartment in MDS-L cells after 7-day treatment with LEN (1 μM). c, Percentage of Annexin V+ cells in the CD34+ and CD34− compartments in MDS-L cells after 7 days of treatment (n = 3 independent experiments). Statistical analysis was performed using two-sided Student’s t-test. Data is presented as mean ± s.d. Source data are available online.
Extended Data Fig. 6 IKZF1 inhibition drives LEN-induced megakaryocytic differentiation in del(5q) cells.
a, Western blot showing efficient degradation of IKZF1 after treatment with the different IMiD compounds (1 µM) for 24h in MDS-L cells. CSNK1A1 is only degraded in the presence of LEN. GAPDH is shown as loading control. (n = 3 independent experiments) b, Flow panels show representative CD41 and CD61 expression of MDS-L (top) or KG-1a (bottom) cells treated with IMIDs for 3 days. c, Western blot showing expression of IKZF1 before and after 24h LEN (1 μM) exposure in control (EGFP) and IKZF1-overexpressing cells. (n = 3 independent experiments). d,e, Methylcellulose colony counts (d) and percentage of apoptosis (e) in IKZF1 WT or EGFP expressing MDS-L cells in the presence of LEN (1 µM) or DMSO as control (n = 3 independent experiments). f, Colony counts from IKZF1 WT or EGFP expressing KG-1a cells after plating in the presence of LEN or DMSO as control (n = 3 independent experiments). g, Western blot showing enforced expression of CSNK1A1. EGFP expression is used as control (n = 3 independent experiments). h, Percentage of apoptosis in MDS-L cells expressing CSNK1A1 or EGFP as control and treated with LEN or DMSO as control for 3 days (n = 3 independent experiments). i, Western blot showing enforced expression of IKZF1-DN. EGFP expression is used as control (n = 3 independent experiments). j–l, Percentage of CD41+/CD61+ cells (j), methylcellulose colony counts (k) and percentage of apoptotic cells (l) from MDS-L cells expressing either EGFP or IKZF1-DN isoform after treatment with LEN (1 µM) or DMSO (n = 3 independent experiments). Statistical analysis was performed using ANOVA with multiple correction adjustment. Data is presented as mean ± s.d. Source data are available online.
Extended Data Fig. 7 TP53 regulates LEN-dependent RUNX1 activation.
a, mRNA transcript levels of the indicated genes in control or TP53-KO cells exposed to LEN (1µM) or DMSO for 72h (n = 3 independent experiments). b, RUNX1 mRNA transcript levels in control and TP53-KO cells after 24h treatment with LEN (1µM) or DMSO (n = 3 independent experiments). c, Representative Western blot showing that induction of RUNX1 protein levels is impaired in TP53-KO cells compared to control cells. GAPDH is shown as loading control. Quantification of RUNX1 protein levels relative to GAPDH are presented for each condition (n = 2 independent experiments). d, CSF1R mRNA transcript levels in control and TP53-KO cells after 24h treatment with LEN (1µM) or DMSO (n = 3 independent experiments). Statistical analysis was performed using ANOVA with multiple correction adjustment (a,d) and Student’s t-test (b). Data is presented as mean ± s.d. Source data are available online.
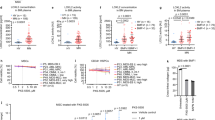
Extended Data Fig. 8 GATA2 drives LEN-induced megakaryocytic differentiation.
e, MDS-L transduced with lentivirus encoding shRNAs targeting GATA2 expression show reduced GATA2 protein levels by Western blot compared to control shRNA-expressing cells. GAPDH is shown as loading control. (n = 3 independent experiments). f, Human ITGA2B (CD41) promoter activation by GATA2 is inhibited by co-expression of IKZF1 in HEK293T cells. (n = 6 independent experiments). Statistical analysis was performed using two-sided Student’s t-test. Data is presented as mean ± s.d. Source data are available online.
Extended Data Fig. 9 RUNX1, GATA2 and IKZF1 bind common genes in human hematopoietic cells.
a, Venn Diagram showing number of common genes bound by IKZF1 and RUNX1/GATA2 in human CD34+ HSPC (left) and K562 cells (right). b, Venn diagram showing intersection of IKZF1/RUNX1/GATA2 ChIP-seq peaks with TP53 ChIP-seq peaks. c, Absence of megakaryocytic or platelet-related pathways in the 167 IKZF1/RUNX1/GATA2 genes co-regulated by TP53 compared to IKZF1/RUNX1/GATA2 gene promoters not bound by TP53. Red dots denote platelet/megakaryocytic-related pathways significantly enriched in each geneset. d, ChIP-seq peak plots obtained from UCSC Genome browser show binding of IKZF1, GATA2, RUNX1 but not TP53 at megakaryocyte-related genes. Black bars denote representation of the available bedfiles for binding of IKZF1 (GSE91730), RUNX1/GATA2 (BloodChIP31) and TP53. Statistical analysis were performed using a chi square test with Yates continuity correction (a,b) or calculated from the Enrichr webserver.
Extended Data Fig. 10 CSNK1A1 haploinsufficiency sensitizes human CD34+ cells to LEN.
Human cord blood CD34+ cells were transduced with shRNA targeting CSNK1A1 or empty vector control and treated with LEN (1 µM) or DMSO for 3 days. a, Gating strategy for the analysis of cord blood cells. b, Analysis by flow cytometry showed increased Annexin V+ staining of LEN-treated shCSNK1A1 cells compared to shControl. (n = 6 independent experiments) c–e, Percentage of CD11b+ (c), CD14+ (d) or CD235a+ (e) cells within the apoptotic (Annexin V+) compartment was not altered by treatment with LEN in shCSNK1A1 cells (n = 3 independent experiments). f, Representative flow panel (left) showing apoptosis (Annexin V+ cells) occurring mainly in the CD34- compartment in shCSNK1A1 cells after 3-day treatment with LEN (1 µM) or DMSO. Percentage of Annexin V+ cells (right) in the CD34+ and CD34- compartments in shCSNK1A1 cells after 3 days of LEN (1 µM) or DMSO treatment (n = 3 independent experiments). Statistical analysis was performed using ANOVA with multiple correction adjustment (Bonferroni). Data is presented as mean ± s.d. Source data are available online.
Supplementary information
Supplementary Tables 1–7
Supplementary Table 1: del(5q) MDS discovery cohort. Supplementary Table 2: list of protein coding variants identified by WGS in the discovery cohort of six paired del(5q) MDS patients. Supplementary Table 3: list of protein coding variants identified by targeted resequencing in an independent validation cohort of del(5q) MDS patients. Supplementary Table 4: variants in the human del(5q) cell lines used in the study. Supplementary Table 5: pathway enrichment in LEN-treated MDS-L cells; n = 3 independent biological samples. Statistical analysis was performed using the Enrichr webserver. Supplementary Table 6: predicted transcription factor activity in MDS-L cells after exposure to LEN for 24 h; n = 3 independent biological samples. Statistical analysis was performed using the Enrichr webserver. Supplementary Table 7: primers used for qPCR, ddPCR and ChIP–PCR assays.
Source data
Source Data Fig. 1
Statistical source data Fig. 1.
Source Data Fig. 1
Unprocessed western blots Fig. 1.
Source Data Fig. 2
Statistical source data Fig. 2.
Source Data Fig. 3
Statistical source data Fig. 3.
Source Data Fig. 4
Statistical source data Fig. 4.
Source Data Fig. 4
Unprocessed western blots Fig. 4.
Source Data Fig. 5
Statistical source data Fig. 5.
Source Data Extended Data Fig. 2
Statistical source data Extended Data Fig. 2.
Source Data Extended Data Fig. 2
Unprocessed western blots Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Statistical source data Extended Data Fig. 3.
Source Data Extended Data Fig. 3
Unprocessed western blots Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Statistical source data Extended Data Fig. 4.
Source Data Extended Data Fig. 4
Unprocessed western blots Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data Extended Data Fig. 6.
Source Data Extended Data Fig. 6
Unprocessed western blots Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Statistical source data Extended Data Fig. 7.
Source Data Extended Data Fig. 7
Unprocessed western blots Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Statistical source data Extended Data Fig. 8.
Source Data Extended Data Fig. 8
Unprocessed western blots Extended Data Fig. 8.
Source Data Extended Data Fig. 10
Statistical source data Extended Data Fig. 10.
Rights and permissions
About this article
Cite this article
Martinez-Høyer, S., Deng, Y., Parker, J. et al. Loss of lenalidomide-induced megakaryocytic differentiation leads to therapy resistance in del(5q) myelodysplastic syndrome. Nat Cell Biol 22, 526–533 (2020). https://doi.org/10.1038/s41556-020-0497-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0497-9