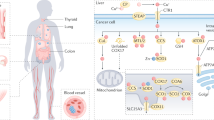
Abstract
Although the transition metal copper (Cu) is an essential nutrient that is conventionally viewed as a static cofactor within enzyme active sites, a non-traditional role for Cu as a modulator of kinase signalling is emerging. Here, we found that Cu is required for the activity of the autophagic kinases ULK1 and ULK2 (ULK1/2) through a direct Cu–ULK1/2 interaction. Genetic loss of the Cu transporter Ctr1 or mutations in ULK1 that disrupt the binding of Cu reduced ULK1/2-dependent signalling and the formation of autophagosome complexes. Increased levels of intracellular Cu are associated with starvation-induced autophagy and are sufficient to enhance ULK1 kinase activity and, in turn, autophagic flux. The growth and survival of lung tumours driven by KRASG12D is diminished in the absence of Ctr1, is dependent on ULK1 Cu binding and is associated with reduced levels of autophagy and signalling. These findings suggest a molecular basis for exploiting Cu-chelation therapy to prevent autophagy signalling to limit proliferation and improve patient survival in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data generated that support the findings of this study are available from the corresponding author on reasonable request. Unprocessed blots have been provided for Figs. 1b–j, 2a,e, 5g, 6a–f and 8a,b, and Extended Data Figs. 1a, 2d,e,g,i,j,l, 6a–c,e,f and 7d. Source data have been provided for Figs. 2b,d,f,h, 3b,g–l, 4f,g,i, 5b,d,f,j, 6e, 7c–e and 8c,e,g, and Extended Data Figs. 1b–k, 2a–c,f,h,k, 3b,e,f, 4b,c, 5b–d,f, 6d,f,h,i and 7a,c,e–g.
References
Kolch, W., Halasz, M., Granovskaya, M. & Kholodenko, B. N. The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer 15, 515–527 (2015).
Festa, R. A. & Thiele, D. J. Copper: an essential metal in biology. Curr. Biol. 21, R877–R883 (2011).
Chelly, J. et al. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet. 3, 14–19 (1993).
Mercer, J. F. B. et al. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat. Genet. 3, 20–25 (1993).
Huster, D. et al. Consequences of copper accumulation in the livers of the Atp7b −/−(Wilson disease gene) knockout mice. Am. J. Pathol. 168, 423–434 (2006).
Pfeiffenberger, J. et al. Hepatobiliary malignancies in Wilson disease. Liver Int. 35, 1615–1622 (2015).
Turski, M. L. et al. A novel role for copper in Ras/MAPK signaling. Mol. Cell. Biol. 32, 1284–1295 (2012).
Brady, D. C. et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509, 496–496 (2014).
Krishnamoorthy, L. et al. Copper regulates cyclic-AMP-dependent lipolysis. Nat. Chem. Biol. 12, 586–592 (2016).
Brady, D. C., Crowe, M. S., Greenberg, D. N. & Counter, C. M. Copper chelation inhibits BRAFV600E-driven melanomagenesis and counters resistance to BRAFV600E and MEK1/2 inhibitors. Cancer Res. 77, 6240–6252 (2017).
Xu, M. M., Casio, M., Range, D. E., Sosa, J. A. & Counter, C. M. Copper chelation as targeted therapy in a mouse model of oncogenic BRAF-driven papillary thyroid cancer. Clin. Cancer Res. 24, 4271–4281 (2018).
Goodman, V. L., Brewer, G. J. & Merajver, S. D. Control of copper status for cancer therapy. Curr. Cancer Drug Targets 5, 543–549 (2005).
Petherick, K. J. et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383 (2015).
Chan, E. Y. W., Kir, S. & Tooze, S. A. siRNA screening of the Kinome Identifies ULK1 as a Multidomain Modulator of Autophagy. J. Biol. Chem. 282, 25464–25474 (2007).
Ganley, I. G. et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Hosokawa, N. et al. Nutrient-dependent mTORCl association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Jung, C. H. et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Kim, D.-H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Hara, K. et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 (1998).
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K.-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008).
Sancak, Y. et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Inoki, K., Zhu, T. & Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P. & Snyder, S. H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43 (1994).
Brown, E. J. et al. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 369, 756–758 (1994).
Brady, G. F. et al. Regulation of the copper chaperone CCS by XIAP-mediated ubiquitination. Mol. Cell Biol. 30, 1923–1936 (2010).
Xiao, T. et al. Copper regulates rest-activity cycles through the locus coeruleus-norepinephrine system. Nat. Chem. Biol. 14, 655–663 (2018).
Chiang, G. G. & Abraham, R. T. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 280, 25485–25490 (2005).
Brown, E. J. et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377, 441–446 (1995).
Soliman, G. A. et al. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J. Biol. Chem. 285, 7866–7879 (2010).
Kim, J., Kundu, M., Viollet, B. & Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Gump, J. M. et al. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat. Cell. Biol. 16, 47–54 (2014).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Itakura, E. & Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010).
Mercer, T. J., Gubas, A. & Tooze, S. A. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 293, 5386–5395 (2018).
Karanasios, E. et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 126, 5224–5238 (2013).
Egan, D. F. et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 (2015).
Russell, R. C. et al. ULK1 induces autophagy by phosphorylating beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 (2013).
Park, Y. S. et al. AKT-induced PKM2 phosphorylation signals for IGF-1-stimulated cancer cell growth. Oncotarget 7, 48155–48167 (2016).
Dooley, H. C. et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 55, 238–252 (2014).
Eng, K. E., Panas, M. D., Hedestam, G. B. K. & McInerney, G. M. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 6, 634–641 (2010).
Amaravadi, R. K., Kimmelman, A. C. & Debnath, J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9, 1167–1181 (2019).
Strohecker, A. M. et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E–driven lung tumors. Cancer Discov. 3, 1272–1285 (2013).
Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Guo, J. Y. et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 27, 1447–1461 (2013).
Bryant, K. L. et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25, 628–640 (2019).
Kinsey, C. G. et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25, 620–627 (2019).
Lee, C.-S. et al. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl Acad. Sci. USA 116, 4508–4517 (2019).
Guo, J. Y. et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 30, 1704–1717 (2016).
McAlpine, F., Williamson, L. E., Tooze, S. A. & Chan, E. Y. W. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy 9, 361–373 (2013).
Lee, J., Peña, M. M. O., Nose, Y. & Thiele, D. J. Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 277, 4380–4387 (2002).
Nose, Y., Kim, B.-E. & Thiele, D. J. Ctr1 drives intestinal copper absorption and is essential for growth, iron metabolism, and neonatal cardiac function. Cell Metab. 4, 235–244 (2006).
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668 (2001).
Walter, D. M. et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature 568, 423–427 (2019).
N’Diaye, E. N. et al. PLIC proteins or ubiquilins regulate autophagy‐dependent cell survival during nutrient starvation. EMBO Rep. 10, 173–179 (2009).
Fung, C., Lock, R., Gao, S., Salas, E. & Debnath, J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol. Biol. Cell 19, 797–806 (2008).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Hara, T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008).
Walter, D. M. et al. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 77, 1719–1729 (2017).
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Safran, M. et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol. Imaging 2, 297–302 (2003).
Hamad, N. M. et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16, 2045–2057 (2002).
Acknowledgements
We thank D. J. Thiele (Duke University), C. J. Chang (University of California Berkeley), R. K. Amaravadi (University of Pennsylvania) and S. A. Tooze (The Francis Crick Institute) for reagents, and R. K. Amaravadi, I. A. Asangani, L. Busino, C. V. Dang, J. M. Davis, T. P. Gade, K. E. Hamilton, B. Keith, M. E. Murphy, R. Natesan, A. M. O’Reilly, S. W. Ryeom, M. C. Simon, B. Z. Stanger, J. Tobias, N. A. Tripp, A. T. Weerartna, K. E. Wellen and E. S. Witze for technical support, discussions and/or reading the manuscript. This work was supported by the American Cancer Society Postdoctoral Fellowship (131203 PF 17 147 01 CCG, to J.M.P.), NIH grants GM124749 (to D.C.B.), CA193603 (to D.M.F.), CA222503 (to D.M.F.), ES019851 (to M.C.) and CA243294 (T.T.), Pew Scholars Program in Biomedical Science Award no. 50359 (to D.C.B.), and V Foundation Scholar Award 3C59 8ABS 3424 3BDA (to D.C.B.).
Author information
Authors and Affiliations
Contributions
T.T. and J.M.P. contributed equally to this research. T.T. contributed to the study design, generated cell lines and plasmids, prepared samples for ICP-MS, performed western-blot analysis, immunocomplex kinase assays, crystal-violet growth assays, co-immunoprecipitations, qPCR and in vivo xenograft mouse work, analysed data and prepared figures. J.M.P. generated cell lines, performed flow cytometry, immunofluorescence imaging, live-cell imaging, immunohistochemical analysis, western-blot analysis, crystal-violet growth assays and qPCR, and assisted with in vivo KRASG12D mouse tumour model work. A.A.G. maintained in vivo KRASG12D mouse tumour models and performed in vivo KRASG12D mouse tumour model imaging. M.C. generated in vivo KRASG12D mouse model tumours. D.M.F. contributed to the study design, provided expertise about in vivo mouse tumour models and facilitated data analysis. D.C.B. conceived the project, contributed to the study design, generated cell lines, plasmids and recombinant proteins, performed Cu-binding assays, in vitro kinase assays, immunofluorescence imaging and in vivo xenograft mouse research, analysed data, prepared figures and wrote the manuscript. All of the authors read and provided feedback on manuscript and figures.
Corresponding author
Ethics declarations
Competing interests
D.C.B. holds ownership in Merlon Inc. D.C.B. is an inventor on the patent application US 20150017261 entitled “Methods of treating and preventing cancer by disrupting the binding of copper in the MAP kinase pathway”. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
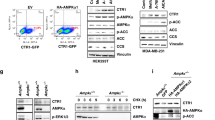
Extended Data Fig. 1 Cu is both necessary and sufficient for ULK1 and ULK2 kinase activity.
a–d, Scatter dot plot with bar at mean normalized ΔPhosphorylated (P)-ATG13/Total (T)-ATG13 from Fig. 1d, e, g. n = 3 biologically independent experiments, Fig. 1d, e, g, or n = 4 biologically independent experiments, Fig. 1f. Results were compared using a one-way ANOVA followed by a Tukey’s multi-comparisons test. 0 vs. 50 μM TTM, *, P = 0.0151; 0 vs. 10 μM MRT68921, *, P = 0.0395. e, RT-PCR detection of indicated mRNAs from Ctr1+/+ MEFs or Ctr1-/- MEFs stably expressing CTR1WT (WT) or empty vector (VO). n = 1 biologically independent sample. f, Scatter dot plot of inductively coupled plasma mass spectrometry (ICP-MS) detection with bar at mean Cu (parts per million, ppm) from Ctr1+/+ MEFs ( + /+) or Ctr1-/- MEFs stably expressing WT or VO per sample weight ± s.e.m. n = 3 biologically independent samples. Results were compared using a one-way ANOVA followed by a Tukey’s multi-comparisons test. *, P = 0.05 0.0202; **, P = 0.0059. g, Representative live cell imaging of the Cu probe CF4 or control Cu probe Ctrl-CF4 from Ctr1+/+ MEFs ( + /+) or Ctr1-/- MEFs stably expressing WT or VO. Scale bar, 100 μm. h, Scatter dot plot with bar at mean CF4 or Ctrl-CF4 fluorescence intensity (FI) ± s.e.m. from Ctr1+/+ MEFs ( + /+) or Ctr1-/- MEFs stably expressing WT or VO. n = 90 individual cells. Results were compared using a one-way ANOVA followed by a Tukey’s multi-comparisons test. ****, P < 0.0001. i–k, Scatter dot plot with bar at mean normalized ΔP-ATG13/T-ATG13 from Fig. 1h–j. n = 3 biologically independent experiments. Results were compared using a one-way ANOVA followed by a Tukey’s multi-comparisons test, an unpaired, two-tailed Student’s t-test, or two-way ANOVA followed by a Sidak’s multi-comparisons test. ns.
Extended Data Fig. 2 Copper is both necessary and sufficient for autophagy induction and signaling in a MAPK signaling independent fashion, upstream of ATG5.
a, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Fig. 2a. n = 3 biologically independent experiments. Results were compared using a two-way ANOVA followed by a Sidak’s multi-comparisons test. ns. b, Scatter dot plot with bar at mean normalized expression of Atp7a mRNA from MEFs n = 1 biologically independent experiment performed in technical triplicate. c, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Fig. 2e. n = 4 biologically independent experiments. Results were compared using a two-way ANOVA followed by a Sidak’s multi-comparisons test. ns. d, Immunoblot detection of proteins from MEFs. e, Immunoblot detection of proteins from treated MEFs. Quantification: ΔLC3-II/β-ACTIN normalized to fl/fl, empty vector (-), VEH control. f, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Extended Data Fig 2e. n = 3 biologically independent experiments. Results were compared using a two-way ANOVA followed by a Tukey’s multi-comparisons test. ns. g, Immunoblot detection of proteins from treated MEFs. Quantification: ΔLC3-II/β-ACTIN normalized to WT, VEH control. h, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Extended Data Fig 2g. n = 6 biologically independent experiments. Results were compared using a two-way ANOVA followed by a Tukey’s multi-comparisons test. ns. i, Immunoblot detection of proteins from MEFs. j, Immunoblot detection of proteins from treated MEFs. Quantification: ΔLC3-II/β-ACTIN normalized to WT, VEH control. k, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Extended Data Fig 2j. n = 5 biologically independent experiments. Results were compared using a two-way ANOVA followed by a Tukey’s multi-comparisons test. *, P = 0.0071; **, P = 0.0434; ***, P = 0.0026; ****, P = 0.0021. l, Immunoblot detection of proteins from treated MEFs. Western blot images are representative of at least three biological replicates.
Extended Data Fig. 3 Cu but not Fe is required for autophagosome formation and is associated with fluctuations in the Cu labile pool.
a, Representative live cell imaging of the Cu probe Ctrl-CF4 every ten minutes for 60 min from MEFs treated with vehicle (VEH) or amino acid deprivation (-AA). Scale bar, 100 μm. b, Mean Ctrl-CF4 fluorescence intensity (FI) ± s.e.m. versus time (minutes, min) from MEFs treated with VEH or -AA normalized to t = 0, five minutes. n = 30 individual cells. Results were compared using a two-way ANOVA followed by a Sidak’s multi-comparisons test. c, Schematic of immunofluorescence-based approach to access autophagosome formation. d, Schematic of flow cytometry-based approach to access autophagosome number. e, f, Scatter dot plot with bar at mean GFP-LC3 fluorescent intensity ± s.e.m. analyzed by flow cytometry from MEFs stably expressing EGFP-LC3B treated VEH or increasing concentrations of Cu chelator TTM (e) or Fe chelator DFO (f). n = 9 biologically independent samples. Results were compared using a one-way ANOVA followed by a Dunnett’s multi-comparisons test. *, P = 0.0148; ****, P < 0.0001.
Extended Data Fig. 4 Genetic ablation of Ctr1 reduces KrasG12D-driven lung tumorigenesis.
a, Normalized representative images of in vivo luminescence of KrasG12D/+;Trp53flox/flox;Rosa26::LSL-Luc (KPLuc) mice introduced with either sgRNA against Glb1 or Ctr1 at week 16 endpoint. b, c, Scatter dot plot with bar at mean ΔLuminescence units from week 10 to week 16 from KPLuc mice introduced with either sgRNA against Glb1 or Ctr1. n = 6 biologically independent animals. Results were compared using an unpaired, one-tailed Student’s t-test. P = 0.0818.
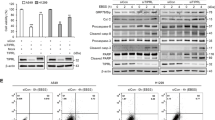
Extended Data Fig. 5 Genetic ablation of Ctr1 decreases MAPK signaling to reduce KrasG12D-driven lung tumorigenesis, while survival in response starvation is independent of MAPK signaling.
a, Representative 40x images of immunohistochemical detection of phosphorylated (P)-ERK1/2 of lungs from KPLuc mice expressing sgRNA against Glb1 or Ctr1. (40x scale bar: 50 µm). b, Scatter dot plot with bar at mean ± s.e.m. % P-ERK1/2 positive staining per tumor (Glb1 and Ctr1) from KPLuc mice expressing sgRNA against Glb1 or Ctr1. n = 66 images. Results were compared using an unpaired, two-tailed Student’s t-test. ***, P = 0.0002. c, Scatter dot plot with bar at mean normalized quantitative PCR (qPCR) expression of Ctr1 mRNA from KP lung adenocarcinoma cell lines #54 (KP #54) and #2474 (KP #2474) stably expressing sgRNA against Rosa (-) or Ctr1 (#1 or #2). n = 1 biologically independent experiment performed in technical triplicate. d, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN from Fig. 5g. n = 3 independent experiments. Results were compared using a two-way ANOVA followed by a Tukey’s multi-comparisons test. ns. e, Representative crystal violet images of KP #54 and KP #2474 cells stably expressing sgRNA against Rosa, Atg5, or Ctr1 (#1 or #2) and ERK2GOF from days 3, 4, and 5 of recovery. f, Scatter dot plot with bar at mean absorbance of extracted crystal violet at 590 nM ± s.e.m. of KP #54 and KP #2474 cells stably expressing sgRNA against Rosa, Atg5, or Ctr1 (#1 or #2) and gain-of-function (GOF) HA-ERK2 (ERK2GOF) from days 3, 4, and 5 of recovery normalized to Rosa, day 3 control. KP #54, n represents number of biologically independent samples; Day 3 n = 9, Day 4 n = 9, Day 5, Rosa n = 8, Atg5 n = 9, #1 n = 9, #2 n = 9. *, P = 0.0140; ***, P = 0.0003. KP #2474, n represents number of biologically independent samples; Day 3 n = 9, Day 4 n = 9, Day 5, Rosa n = 9, Atg5 n = 9, #1 n = 8, #2 n = 8. *, P = 0.0011; **, P = 0.0158; ***, P = 0.0462.
Extended Data Fig. 6 Binding of Cu to ULK1 is required for kinase activity but not substrate association or phosphorylation.
a, Immunoblot detection of phosphorylated (P) ATG13, total (T)- ATG13, P-ULK1 (S555), P-ULK1 (S757), T-ULK1, or β-ACTIN from Ulk1/2-/- MEFs stably expressing HA-ULK1WT (WT) or HA-ULK1CBM (CBM) treated with vehicle (VEH) or amino acid deprivation (-AA). b, Immunoblot detection of T-ULK1 or β-ACTIN from cell lysates treated with or without calf alkaline phosphatase (CIP) from Ulk1/2-/- MEFs stably expressing empty vector (VO), HA-ULK1WT (WT), or HA-ULK1CBM (CBM). c, Immunoblot detection of T-ATG13, T-ATG101, T-FIP200, T-ULK1, or β-ACTIN from immunoprecipitated (IP)-ULK1 or whole cell extracts (WCE) from Ulk1/2-/- MEFs stably expressing HA-ULK1WT (WT) or HA-ULK1CBM (CBM) treated with VEH or -AA. d, Scatter dot plot with bar at mean normalized quantitative PCR (qPCR) expression of Ulk1 or Ulk2 mRNA from MEFs stably expressing sgRNA against Rosa or Ulk1 and Ulk2. n = 1 biologically independent experiment performed in technical triplicate. e, Immunoblot detection of T-ULK1, T-ULK2, or β-ACTIN from MEFs stably expressing sgRNA against Rosa (-) or Ulk1 and Ulk2 ( + ). f, Immunoblot detection of LC3-I, LC3-II, or β-ACTIN from MEFs stably expressing sgRNA against Rosa (-) or Ulk1 and Ulk2 ( + ) treated with -AA and bafilomycin (BAF) for 1 h (hr), 2 hr, and 3 hr. g–i, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN or ΔP-ATG13/T-ATG13 from Fig. 6d, f. n = 3 independent experiments. Results were compared using a two-way ANOVA followed by a Tukey’s multi-comparisons test. g, ns; h, ****, P < 0.0001. i, *, P = 0.0365; ****, P < 0.0001. Western blot images are representative of at least three biological replicates.
Extended Data Fig. 7 Binding of Cu to ULK1 is required for tumorigenesis by oncogenic KRASG12D.
a, Mean tumor volume (mm3) ± s.e.m. versus time (days) in mice injected with Ulk1-/- MEFs stably expressing either HA-ULK1WT or HA-ULK1CBM and transformed with KRASG12D. n = 4 biologically independent animals. Results were compared using a paired, one-tailed Student’s t-test. **, P = 0.0095. b, Representative dissected tumors from mice injected with Ulk1-/- MEFs stably expressing either HA-ULK1WT (WT) or HA-ULK1CBM (CBM) and transformed with KRASG12D. Scale bar, 100 µm. c, Scatter dot plot with bar at mean tumor weight (g) ± s.e.m. of tumors at endpoint from Ulk1-/- MEFs stably expressing either WT or CBM and transformed with KRASG12D. n = 4 biologically independent samples. Results were compared using an unpaired, one-tailed Student’s t-test. *, P = 0.0325. n = 4. d, Immunoblot detection of T-ULK1, T-ULK2, or β-ACTIN from KrasG12D/+;Trp53flox/flox (KP) lung adenocarcinoma cell line #2474 (KP #2474) stably expressing sgRNA against Rosa (-) or Ulk1 and Ulk2 ( + ). e, f, Scatter dot plot with bar at mean normalized ΔLC3-II/β-ACTIN or ΔP-ATG13/T-ATG13 from Fig. 8a, b. n = 3 independent experiments. Results were compared using a one-way ANOVA or a two-way ANOVA followed by a Tukey’s multi-comparisons test. e, **, P = 0.0013; ***, P = 0.0016; ****, P < 0.0001; f, *, P = 0.0113; **, P = 0.0063; ***, P = 0.0113. g, Scatter dot plot with bar at mean normalized quantitative PCR (qPCR) expression of Ctr1 mRNA from KP #2474 cells stably expressing sgRNA against Rosa, Ctr1 #2, or Ulk1, Ulk2, and Ctr1 #2. n = 1 biologically independent experiment performed in technical triplicate.
Supplementary information
Supplementary Information
Supplementary Fig. 1: an example of the gating strategy for the FACS experiments.
Supplementary Video 1
Representative live-cell imaging of the Cu probe CF4 every 10 min for 60 min from MEFs treated with vehicle.
Supplementary Video 2
Representative live-cell imaging of the Cu probe CF4 every 10 min for 60 min from MEFs treated with amino acid deprivation.
Supplementary Video 3
Representative live-cell imaging of the Cu probe Ctrl-CF4 every 10 min for 60 min from MEFs treated with vehicle.
Supplementary Video 4
Representative live-cell imaging of the Cu probe Ctrl-CF4 every 10 min for 60 min from MEFs treated with amino acid deprivation.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 2
Statistical source data
Source Data Fig. 2
Unprocessed western blots
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 5
Unprocessed western blots
Source Data Fig. 6
Statistical source data
Source Data Fig. 6
Unprocessed western blots
Source Data Fig. 7
Statistical source data
Source Data Fig. 8
Statistical source data
Source Data Fig. 8
Unprocessed western blots
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 1
Unprocessed western blots/ or gels
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 2
Unprocessed western blots
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 6
Unprocessed western blots
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 7
Unprocessed western blots
Rights and permissions
About this article
Cite this article
Tsang, T., Posimo, J.M., Gudiel, A.A. et al. Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma. Nat Cell Biol 22, 412–424 (2020). https://doi.org/10.1038/s41556-020-0481-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0481-4
This article is cited by
-
Targeting cuproplasia and cuproptosis in cancer
Nature Reviews Clinical Oncology (2024)
-
Cuprotosis clusters predict prognosis and immunotherapy response in low-grade glioma
Apoptosis (2024)
-
Heavy metals in biological samples of cancer patients: a systematic literature review
BioMetals (2024)
-
Cuproptosis: mechanisms and links with cancers
Molecular Cancer (2023)
-
Iron and copper: critical executioners of ferroptosis, cuproptosis and other forms of cell death
Cell Communication and Signaling (2023)