Abstract
Extra-embryonic mesoderm (ExM)—composed of the earliest cells that traverse the primitive streak—gives rise to the endothelium as well as haematopoietic progenitors in the developing yolk sac. How a specific subset of ExM becomes committed to a haematopoietic fate remains unclear. Here we demonstrate using an embryonic stem cell model that transient expression of the T-box transcription factor Eomesodermin (Eomes) governs haemogenic competency of ExM. Eomes regulates the accessibility of enhancers that the transcription factor stem cell leukaemia (SCL) normally utilizes to specify primitive erythrocytes and is essential for the normal development of Runx1+ haemogenic endothelium. Single-cell RNA sequencing suggests that Eomes loss of function profoundly blocks the formation of blood progenitors but not specification of Flk-1+ haematoendothelial progenitors. Our findings place Eomes at the top of the transcriptional hierarchy regulating early blood formation and suggest that haemogenic competence is endowed earlier during embryonic development than was previously appreciated.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq, scRNA-seq, ChIP-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus (GSE140005). Previously published sequencing data that were re-analysed here are available under accession codes GSE110164, GSE128466 and GSE47085. Source data are provided with this paper. All other data supporting the findings of this study and biological materials presented in this study are available upon reasonable request.
Code availability
All of the computational code is available from E.J.R. upon reasonable request.
Change history
12 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41556-021-00645-6
References
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Kinder, S. J. et al. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126, 4691–4701 (1999).
Ferkowicz, M. J. & Yoder, M. C. Blood island formation: longstanding observations and modern interpretations. Exp. Hematol. 33, 1041–1047 (2005).
McGrath, K. E., Frame, J. M. & Palis, J. Early hematopoiesis and macrophage development. Semin. Immunol. 27, 379–387 (2015).
Frame, J. M., Fegan, K. H., Conway, S. J., McGrath, K. E. & Palis, J. Definitive hematopoiesis in the yolk sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 34, 431–444 (2016).
Kissa, K. & Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010).
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010).
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010).
McGrath, K. E. et al. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11, 1892–1904 (2015).
De Bruijn, M. F., Speck, N. A., Peeters, M. C. & Dzierzak, E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474 (2000).
Keller, G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 19, 1129–1155 (2005).
Murry, C. E. & Keller, G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 (2008).
Porcher, C., Chagraoui, H. & Kristiansen, M. S. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood 129, 2051–2060 (2017).
Lancrin, C. et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457, 892–895 (2009).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Porcher, C. et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86, 47–57 (1996).
Shivdasani, R. A., Mayer, E. L. & Orkin, S. H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434 (1995).
De Bruijn, M. & Dzierzak, E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 129, 2061–2069 (2017).
Huber, T. L., Kouskoff, V., Fehling, H. J., Palis, J. & Keller, G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–630 (2004).
Lugus, J. J., Park, C., Ma, Y. D. & Choi, K. Both primitive and definitive blood cells are derived from Flk-1+ mesoderm. Blood 113, 563–566 (2009).
Arnold, S. J., Sugnaseelan, J., Groszer, M., Srinivas, S. & Robertson, E. J. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis 47, 775–781 (2009).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Arnold, S. J., Hofmann, U. K., Bikoff, E. K. & Robertson, E. J. Pivotal roles for Eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501–511 (2008).
Shalaby, F. et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 (1995).
Lee, D. et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507 (2008).
Koyano-Nakagawa, N. & Garry, D. J. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc. Res. 113, 1294–1306 (2017).
Nostro, M. C., Cheng, X., Keller, G. M. & Gadue, P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 (2008).
Irion, S. et al. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development 137, 2829–2839 (2010).
Chagraoui, H. et al. SCL/TAL1 cooperates with Polycomb RYBP-PRC1 to suppress alternative lineages in blood-fated cells. Nat. Commun. 9, 5375 (2018).
Mikkola, H. K., Fujiwara, Y., Schlaeger, T. M., Traver, D. & Orkin, S. H. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood 101, 508–516 (2003).
Kattman, S. J. et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240 (2011).
Izumi, N., Era, T., Akimaru, H., Yasunaga, M. & Nishikawa, S. Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25, 1664–1674 (2007).
Mitjavila-Garcia, M. T. et al. Expression of CD41 on hematopoietic progenitors derived from embryonic hematopoietic cells. Development 129, 2003–2013 (2002).
Ferkowicz, M. J. et al. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development 130, 4393–4403 (2003).
Costello, I. et al. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084–1091 (2011).
Van Den Ameele, J. et al. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 13, 355–362 (2012).
Clarke, R. L. et al. The expression of Sox17 identifies and regulates hemogenic endothelium. Nat. Cell Biol. 15, 502–510 (2013).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Org, T. et al. Scl binds to primed enhancers in mesoderm to regulate hematopoietic and cardiac fate divergence. EMBO J. 34, 759–777 (2015).
Schutte, J. et al. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife 5, e11469 (2016).
Onodera, K. et al. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl Acad. Sci. USA 94, 4487–4492 (1997).
Teo, A. K. K. et al. Pluripotency factors regulate definitive endoderm specification through Eomesodermin. Genes Dev. 25, 238–250 (2011).
Vijayaragavan, K. et al. Noncanonical Wnt signaling orchestrates early developmental events toward hematopoietic cell fate from human embryonic stem cells. Cell Stem Cell 4, 248–262 (2009).
Ng, E. S. et al. The primitive streak gene Mixl1 is required for efficient haematopoiesis and BMP4-induced ventral mesoderm patterning in differentiating ES cells. Development 132, 873–884 (2005).
DiMartino, J. F. et al. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98, 618–626 (2001).
Fleury, M., Eliades, A., Carlsson, P., Lacaud, G. & Kouskoff, V. FOXF1 inhibits hematopoietic lineage commitment during early mesoderm specification. Development 142, 3307–3320 (2015).
Wang, M. et al. MEIS2 regulates endothelial to hematopoietic transition of human embryonic stem cells by targeting TAL1. Stem Cell Res. Ther. 9, 340 (2018).
Davenport, T. G., Jerome-Majewska, L. A. & Papaioannou, V. E. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development 130, 2263–2273 (2003).
Goode, D. K. et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell 36, 572–587 (2016).
Wang, Q. et al. The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20, 70–86 (2017).
Tosic, J. et al. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 21, 1518–1531 (2019).
Lie, A. L. M. et al. Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development 145, dev149419 (2018).
Fitch, S. R. et al. Gata3 targets Runx1 in the embryonic haematopoietic stem cell niche. IUBMB Life 72, 45–52 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Eliades, A. et al. The hemogenic competence of endothelial progenitors is restricted by Runx1 silencing during embryonic development. Cell Rep. 15, 2185–2199 (2016).
Tober, J., Yzaguirre, A. D., Piwarzyk, E. & Speck, N. A. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765–3776 (2013).
Yzaguirre, A. D., Howell, E. D., Li, Y., Liu, Z. & Speck, N. A. Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis. Development 145, dev158162 (2018).
Swiers, G. et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, 2924 (2013).
Obier, N. et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development 143, 4324–4340 (2016).
Padron-Barthe, L. et al. Clonal analysis identifies hemogenic endothelium as the source of the blood–endothelial common lineage in the mouse embryo. Blood 124, 2523–2532 (2014).
Behringer, R., Gertsenstein, M., Nagy, K. V. & Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2014).
DiTacchio, L. et al. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J. Biol. Chem. 287, 23657–23666 (2012).
Wang, Q. et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA 93, 3444–3449 (1996).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
McLeay, R. C. & Bailey, T. L. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinf. 11, 165 (2010).
Zhao, H. et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Robinson, J. T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Stemmer, M., Thumberger, T., Del Sol Keyer, M., Wittbrodt, J. & Mateo, J. L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10, e0124633 (2015).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Senft, A. D. et al. Combinatorial Smad2/3 activities downstream of nodal signaling maintain embryonic/extra-embryonic cell identities during lineage priming. Cell Rep. 24, 1977–1985.e7 (2018).
Acknowledgements
We acknowledge M. Maj and L. Ericsen (flow cytometry facility at the Dunn School) and K. Clark (flow cytometry facility at the Weatherall Institute of Molecular Medicine (WIMM)) for providing cell sorting services. The WIMM is supported by the MRC HIU, MRC MHU (MC_UU_12009), NIHR Oxford BRC and John Fell Fund (131/030 and 101/517), EPA fund (CF182 and CF170) and WIMM Strategic Alliance awards G0902418 and MC_UU_12025. We thank N. Ashley for help with 10× sample preparation and sequencing. The WIMM Single Cell Core Facility was supported by the MRC MHU (MC_UU_12009), the Oxford Single Cell Biology Consortium (MR/M00919X/1) and WT-ISSF (097813/Z/11/B#) funding. The WIMM facility was supported by WIMM Strategic Alliance awards G0902418 and MC_UU_12025. We also thank the High-Throughput Genomics Group (Wellcome Trust Centre for Human Genetics, funded by the Wellcome Trust (090532/Z/09/Z)) for generating sequencing data. We thank V. Kouskoff for providing the iRunx1 embryonic stem cell line, S. Thongjuea and G. Wang for advice on the scRNA-seq analysis, J. Riepsaame for advice on the CRISPR experiments, and D. Higgs, H. Chagraoui, D. Owens, A. Nelson and A. Mould for helpful discussions. M.F.T.R.d.B. and C.P. are supported by programmes in the MRC Molecular Hematology Unit Core award (grant number MC_UU_12009/2 to M.F.T.R.d.B. and MC_UU_12009/9 to C.P.). L.G. was supported by a Clarendon PhD studentship and the MRC Molecular Haematology Unit. The work was supported by grants from the Wellcome Trust (214175/Z/18/Z to E.J.R. and 10281/Z/13/Z to L.T.G.H.). L.T.G.H. was supported by a Clarendon Fund Scholarship and Trinity College Titley Scholarship. E.J.R. is a Wellcome Trust Principal Fellow.
Author information
Authors and Affiliations
Contributions
L.T.G.H., E.K.B., C.P., M.F.T.R.d.B. and E.J.R. designed the study. L.T.G.H., C.S.S., I.C. and A.D.S. performed the experiments. L.T.G.H., I.I.-R., J.C.M. and B.G. performed the scRNA-seq analyses. L.G. generated the Runx1-Venus reporter line. L.T.G.H., E.K.B., M.F.T.R.d.B. and E.J.R. wrote the manuscript with input from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Valerie Kouskoff and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
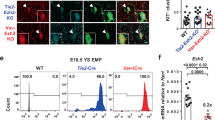
Extended Data Fig. 1 Eomes and Flk-1 expression at the outset of mouse gastrulation.
a,b, Immunofluorescence staining of E6.5 (a) and E7.5 (b) wildtype embryos for Flk-1 (red) and Eomes (green); Images representative of 5 embryos (E6.5) and 8 embryos (E7.5). Nuclei are stained with DAPI (blue). Dotted white lines indicate the extraembryonic/embryonic boundary. White boxes denote the zoomed in areas displayed in the far-right panel. White arrows indicate Flk-1+ extra-embryonic mesodermal cells. Orange arrows indicate Flk-1+ embryonic mesodermal cells. Scale bars, 100 μM.
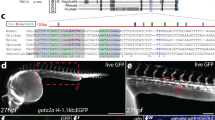
Extended Data Fig. 2 Generation of the EomesiCre allele.
a, Targeting strategy used to generate the EomesiCre allele. The targeting vector introduces a mammalian codon-improved Cre (iCre), β-globin polyA cassette into the endogenous Eomes initiator methionine and a LoxP flanked PGK-Neo positive drug selection cassette and TK negative selection cassette. Red and blue lines indicate the locations of 5’ external (red) and 3’ external probes (blue, exon 6) used for Southern blotting. Orange arrows indicate the location of primers used for PCR genotyping. E=EcoRV. b, Southern blot showing wildtype (15 kb) and targeted (10 kb) alleles. (Red probe) c, Southern blot after Cre-mediated excision of the Neo drug selection cassette showing excised targeted (6 kb) and wild type (15 kb) alleles and loss of the targeted allele (8 kb). (Blue probe) d, PCR genotyping of EomesiCre mice; iCre band size is 440 bp. (b-d) Southern blots and PCR genotyping blots are representative of at least 3 independent experiments. e, Wholemount in-situ hybridization (WISH) comparing Eomes and iCre expression in EomesiCre/+ embryos. ExE, extra-embryonic ectoderm; ch, chorion; PS, primitive streak; APS, anterior primitive streak. Scale bars, 100 μM f, Evaluating the expression of extra-embryonic mesodermal markers upon Eomes conditional inactivation from the epiblast (EomesΔepi). (i) Wholemount X-gal staining of E7.5 control (top) and mutant EomesΔepi (bottom) embryos carrying a Flk-1-LacZ reporter allele. (ii) WISH analysis of ER71 expression in control (top) and mutant EomesΔepi (bottom) embryos. Scale bars, 100 μm. LS, late streak; EB, early allantoic bud stage. (e-f) Images are representative of at least 3 embryos from each gestational stage and genotype.
Extended Data Fig. 3 Generation of the Runx1-Venus ESC reporter line.
a, Targeting strategy used to generate the Runx1-Venus allele. A 3xFlag-P2A-Venus was inserted in the last exon of Runx1 (exon 6), before the stop codon. Pink arrow indicates the locations of Cas9-gRNA induced DNA double stand break immediately preceding the Runx1 stop codon. Orange arrows indicate locations of primers used for PCR genotyping b, Integration of the targeting vector was confirmed using long-range PCR; Runx-1Venus band size is 5.8 kb, WT band size is 4.4 kb. A clone with the Venus reporter integrated in both alleles (RV11 – red box) was used for all experiments. This experiment was performed once. c, Assessing Eomes’ requirement for cell autonomous Runx1 expression during hematopoiesis (left panel). At day 0 WT (grey) cells were mixed in equal proportions with either Eo+/+ Runx1-Venus ESCs (orange) or Eo-/- Runx1-Venus ESCs (purple). Flow cytometric analysis was performed on bulk EHT RV-Eo+/+:WT (top) and RV-Eo-/-:WT co-cultures at day 6 to assess Runx1-venus, cKit and CD41 expression. Runx1-venus was expressed at varying levels (left flow cytometry panel); Runx1-negative, grey dots; Runx1-lo, blue dots; Runx1 + , yellow dots. Percentages of Runx1-Venuslo (blue) and Runx1-Venus+ (yellow) cells within the CD41hi cell population are indicated by coloured numbers in the middle and right flow cytometry panels; Representative of 1 Runx-1 Venus Eomes+/+ and 2 Runx1-Venus Eomes-/- clones.
Extended Data Fig. 4 Re-generation of the Eomes loss-of-function allele in the SCL-mCherry and Runx1-Venus ESC reporter lines and the iRunx1 Runx1-/- ESC using CRISPR-Cas9.
a, The targeting strategy used to re-generate the Eomes-loss of function allele23 in SCL-mCherry and Runx1-Venus ESC reporter lines. An ssODN was used to patch an EcoRV site in between introns 1 and 5 to delete a 3.5 kb region, including exons 2-5 of the Eomes locus. Pink arrows indicate the locations of Cas9-gRNA induced DNA double strand breaks in intron 1 and intron 5. Blue lines (exon 6) indicate the location of a 3’ external Southern blotting probe. Orange arrows indicate the location of primers used for PCR genotyping. E = EcoRV. b, Southern blots showing wild type (15 kb) and targeted (2.5 kb) alleles in Runx1-Venus (top) and SCL-mCherry (bottom) ESC lines. Representative of 2 independent experiments. c, PCR genotyping of Eomes-null ESCs; WT band size is 401 bp, Eomes-null band size is 535 bp. d, The targeting strategy used to re-generate the Eomes-loss of function allele in the iRunx1 Runx1-/- ESC line. ssODNs were used to patch an Sph1/Spe1 restriction sites in between introns 1 and 5 to delete a 3.5 kb region, including exons 2-5 of the Eomes locus. Pink arrows indicate the locations of Cas9-gRNA induced DNA double strand breaks in intron 1 and intron 5. Orange arrows indicate the location of primers used for PCR genotyping. e, PCR genotyping of Eomes-null ESCs; WT band size is 499 bp, Eomes-null band size is 633 bp and digestion products of the Eomes-null PCR product are 221 bp and 412 bp. iRunx1 Runx1-/- Eomes-/- clones had insertion of both Sph1 and Spe1 restriction sites, indicating a homozygous deletion. Representative of 2 independent experiments.
Extended Data Fig. 5 Generation of the EomesV5/V5 ESC line for ChIP-Seq.
a, The targeting strategy used to insert a 3XGly-V5 tag directly upstream of the UAG translational stop codon (exon 6) at the C-terminus of the Eomes locus using an ssODN. The pink arrow indicates the location of the Cas9-gRNA induced DNA double strand break. Orange arrows indicate the location of primers used for PCR genotyping. b, Western blotting of whole cell protein lysates from EoV5/V5 and WT day 4 EBs shows that the three isoforms of Eomes protein (upper panel) are V5 tagged (lower panel) in both EoV5/V5 clones. This experiment was performed once. c, PCR genotyping of Eomes-V5 targeted clones; WT band size is 250 bp, Eo-V5 band size is 301 bp. Representative of at least 3 independent experiments. d, e, V5 protein expression was evaluated in day 3-5 EomesV5/V5 EBs using intracellular flow cytometry. d Equivalent stage WT EBs were used as a gating control (upper panel). e, Additionally, EBs were stained for Flk-1 and PdgfRa expression; purple dots indicate cells that are V5 + and numbers indicate the proportion of cells in the Flk-1hi/PdgfRa- that are V5+; Representative of 1 wildtype and 2 EoV5/V5 clones. f, Representative immunofluorescence staining of EoV5/V5 day 4 EBs for Eomes (red) and V5 (green); 2 biologically independent samples. Nuclei are counter stained with DAPI (blue). Scale bars, 100 μM. g, Representative flow cytometric analysis of Flk-1/PdgfRa (top) and CD41/c-Kit (bottom) expression in WT (left) and EoV5/V5 (right) day 4 and day 7 EBs, respectively; Representative of1 WT and 2 EoV5/V5 clones.
Extended Data Fig. 6 Eomes binds multiple Runx1 cis-regulatory regions.
IGV snapshots of EomesV5/V5, Eomes-V5/ Eomes-GFP51, Tead449, Smad2/350 and Scl39 ChIP-Seq peaks. H3K27Ac ChIP-Seq peaks and DNase1 hypersensitivity (DN1 HS) at different stages of hematopoietic development in vitro; HB, hemangioblast (T+ /Flk-1+); HE, hemogenic endothelium (Tie2+/Kit+); HP, hematopoietic progenitor (CD41+)49. Yellow bars highlight Eomes bound sites identified in the Eomes-V5 dataset51. The numbers above coloured columns indicate the relative location of these sites in kilobases to the TSS at P1 of the Runx1 locus. Red numbers indicate sites which have reduced chromatin accessibility in Eomes-/- Flk-1hi/PdgfRa- cells. P1, promoter 1; P2, promoter 2.
Extended Data Fig. 7 Comparing Eomes bound ChIP-Seq peaks with the subset of ATAC-Seq peaks that are Eomes-dependent and normally bound by SCL at various stages of hematopoietic differentiation.
a, Venn diagram depicting the overlap of Eo-V5 ChIP-Seq peaks in D4 EBs, SCL ChIP-Seq peaks in D4 Flk-1+ EBs39 and sites of reduced chromatin accessibility in day 4 Eo-/- Flk-1+/PdgfRa- cells. Heatmaps showing ATAC signals (blue) from WT or Eomes-/- day 4 Flk-1hi/PdgfRa- cells and SCL ChIP-Seq signal (green) from day 4 WT Flk-1+ cells39. All heatmaps show a 4 kb region flanking peak centres of the 338 Eomes bound sites. b, Venn diagram depicting the overlap of Eo-V5 ChIP-Seq peaks in D4 EBs with sites of reduced chromatin accessibility in day 4 Eo-/- Flk-1hi/PdgfRa- cells that are normally bound by SCL in WT day 4 Flk-1+ cells39. Heatmaps show DNAseI hypersensitivity signal (red), ChIP-Seq signal for H3K27Ac (green) histone modifications and SCL occupancy (purple) in different cell populations that develop during hematopoietic differentiation. Mes, mesoderm (T+/Flk-1-); HB, hemangioblast (T+/Flk-1+); HE, hemogenic endothelium (Tie2+/Kit+); HP, hematopoietic progenitor (CD41+)49. All heatmaps show 4 kb regions flanking peak the peak regions indicated in the Venn diagram.
Extended Data Fig. 8 scRNA-Seq shows that hematoendothelial progenitor and endothelial subsets are still specified in the absence of Eomes function.
a, Gating strategy used to sort Flk1hi/PdgfRa- cells from day 4 and day 5 wildtype and Eomes-/- EBs for 10X scRNA-Seq. b, c, Uniform manifold approximation and projection (UMAP) plots. WT (blue) and Eomes-/- (red) Flk-1hi/PdgfRa- cells from day 4 (left) and day 5 (right) EBs; n = 3805 cells/genotype/day. c, WT (left) and Eomes-/- (right) cells coloured by their Seurat cluster/cell type annotation denoted in the legend. The black box highlights clusters 7 and 8 that are diminished in the Eomes-/- cultures. d, Log2 fold change in the abundance of Eomes-/- cells with respect to WT cells in each Seurat cluster. e, Violin plots showing mesodermal, hematovascular and allantoic gene expression levels in WT (blue) and Eomes-/- (red) cells within the various Seurat clusters; n = 7610 WT cells and n = 7610 Eomes-/- cells. Violin plots show normalized expression values, genes are indicated on the x-axis and clusters on the y-axis. f, Normalized expression of Scl (top) and Runx1 (bottom) overlaid on UMAPs for all single cells collected from day 4/5 Flkhi/PdgfRa- wildtype (left) or Eomes-/- (right) EBs. HEP, hematoendothelial progenitors; Endo, endothelium; Meso, mesoderm; PS, primitive streak.
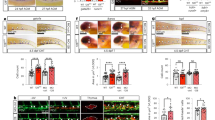
Extended Data Fig. 9 Eomes-null endothelial cells immunophenocopy Runx1-null endothelial cells.
a, b, Flow cytometric analysis of day 6 EB (a) and day 7 EHT (b) for expression of Cdh5/cKit and CD41 within the Cdh5+/cKit+ compartment. Pink gate highlights a CD41lo subset absent in SCL-null cultures. This experiment was performed once. c, Proportion of Cdh5+ cells in day 8 iRunx1 Eomes+/+ and iRunx1 Eomes -/- EHT cultures in which Runx1 has been uninduced (0 ng/mL dox) or induced (90 and 300 ng/mL dox) from day 6-8. Graphical representations depict mean± SEM of n = 3 independent differentiations for iRunx1 Eo+/+ 90 ng/mL and iRunx1 Eo+/+ 300 ng/mL and n = 4 independent differentiations for iRunx1 Eo+/+ 0 ng/mL, iRunx1 Eo-/- 0 ng/mL, iRunx1 Eo-/- 90 ng/mL and iRunx1 Eo-/- 300 ng/mL. Statistical source data are provided in Source Data Fig. 8. d, Representative examples of definitive erythro-myeloid colonies generated from iRunx1 Eomes-/- day 8 EHT cultures in which Runx1 expression was induced from day 6-8 via dox addition; 3 independent differentiations. Ery-D, definitive erythrocyte; GM, granulocyte-macrophage; GEMM, granulocyte-erythrocyte-myeloid. Scale bars, 100 μM.
Supplementary information
Supplementary Tables
Supplementary Table 1. List of conserved marker genes across genotypes for each Seurat cluster. Only positive markers for each cluster are listed. Supplementary Table 2. Primer sequences. Primer sequences used for real-time quantitative reverse transcription PCR and genotyping are listed. Product lengths indicate the size of the PCR product in base pairs. Supplementary Table 3. Antibody information. Antibodies used for immunofluorescence, flow cytometry, ChIP, western blotting and MACS experiments are listed. Supplementary Table 4. iCre probe sequence. The sequence for the iCre probe used for whole-mount in situ hybridization is shown. Supplementary Table 5. CRISPR–Cas9 reagents. Sequences for gRNAs and repair templates are listed that were used to generate the Runx1-Venus, Eomes-V5 and Eomes-null ESC lines. Asterisks indicate phosphorothioated DNA bases. Supplementary Table 6. Eomes-V5-bound genomic regions. List of the Eomes-V5-bound genomic regions and nearby associated genes, as identified using GREAT. Additionally, lists on the right indicate whether the corresponding Eomes-V5-bound peak is co-occupied by Tead4 (ref. 49) and Smad2/3 (ref. 50) in the indicated datasets.
Source data
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed southern blots.
Source Data Extended Data Fig. 4
Unprocessed Southern blots/gels.
Source Data Extended Data Fig. 5
Unprocessed western blots/gels.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Harland, L.T.G., Simon, C.S., Senft, A.D. et al. The T-box transcription factor Eomesodermin governs haemogenic competence of yolk sac mesodermal progenitors. Nat Cell Biol 23, 61–74 (2021). https://doi.org/10.1038/s41556-020-00611-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-00611-8
This article is cited by
-
Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development
Nature Communications (2022)