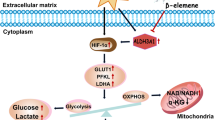
Abstract
Mutant KRAS modulates the metabolic plasticity of cancer cells to confer a growth advantage during hypoxia, but the molecular underpinnings are largely unknown. Using a lipidomic screen, we found that PLCγ1 is suppressed during hypoxia in KRAS-mutant human lung adenocarcinoma cancer cell lines. Suppression of PLCγ1 in hypoxia promotes a less oxidative cancer cell metabolism state, reduces the formation of mitochondrial reactive oxygen species and switches tumour bioenergetics towards glycolysis by impairing Ca2+ entry into the mitochondria. This event prevents lipid peroxidation, antagonizes apoptosis and increases cancer cell proliferation. Accordingly, loss of function of Plcg1 in a mouse model of KrasG12D-driven lung adenocarcinoma increased the expression of glycolytic genes, boosted tumour growth and reduced survival. In patients with KRAS-mutant lung adenocarcinomas, low PLCγ1 expression correlates with increased expression of hypoxia markers and predicts poor patient survival. Thus, our work reveals a mechanism of cancer cell adaptation to hypoxia with potential therapeutic value.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All supporting data are included in this published article and its Supplementary Information. Requests to use the PLCγ1 floxed mice will be redirected to F.H.H. and T.M.S. The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) dataset was retrieved from the Genomic Data Commons Portal: http://cancergenome.nih.gov. The data were downloaded with the help of the web graphic user interface Xena browser https://xenabrowser.net. Source data are provided with this paper.
References
Thomlinson, R. H. & Gray, L. H. The histological structure of some human lung cancers and the possible implications for radiotherapy. Br. J. Cancer 9, 539–549 (1955).
Brown, J. M. & Wilson, W. R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447 (2004).
Bhandari, V. et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 51, 308–318 (2019).
Kamphorst, J. J. et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl Acad. Sci. USA 110, 8882–8887 (2013).
Bensaad, K. et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia–reoxygenation. Cell Rep. 9, 349–365 (2014).
Botto, L. et al. Hypoxia-induced modifications in plasma membranes and lipid microdomains in A549 cells and primary human alveolar cells. J. Cell. Biochem. 105, 503–513 (2008).
Huang et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 8, 1930–1942 (2014).
Semenza, G. L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 (2013).
Listenberger, L. L. et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl Acad. Sci. USA 100, 3077–3082 (2003).
Bailey, A. P. et al. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell 163, 340–353 (2015).
D’Souza, K. & Epand, R. M. Enrichment of phosphatidylinositols with specific acyl chains. Biochim. Biophys. Acta 1838, 1501–1508 (2014).
Wymann, M. P. & Schneiter, R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9, 162–176 (2008).
Balla, T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 (2013).
Saliakoura, M. et al. The ACSL3–LPIAT1 signaling drives prostaglandin synthesis in non-small cell lung cancer. Oncogene 39, 2948–2960 (2020).
Bunney, T. D. & Katan, M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer 10, 342–352 (2010).
Schramp, M., Hedman, A., Li, W., Tan, X. & Anderson, R. PIP kinases from the cell membrane to the nucleus. Subcell. Biochem. 58, 25–59 (2012).
Koss, H., Bunney, T. D., Behjati, S. & Katan, M. Dysfunction of phospholipase Cγ in immune disorders and cancer. Trends Biochem. Sci. 39, 603–611 (2014).
Zhong, H. et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60, 1541–1545 (2000).
Mazure, N. M., Chen, E. Y., Laderoute, K. R. & Giaccia, A. J. Induction of vascular endothelial growth factor by hypoxia is modulated by a phosphatidylinositol 3-kinase/Akt signaling pathway in Ha-ras-transformed cells through a hypoxia inducible factor-1 transcriptional element. Blood 90, 3322–3331 (1997).
Ferrer, I. et al. KRAS-mutant non-small cell lung cancer: from biology to therapy. Lung Cancer 124, 53–64 (2018).
Salem, A. et al. Targeting hypoxia to improve non-small cell lung cancer outcome. J. Natl Cancer Inst. 110, 14–30 (2018).
Gazdar, A. F., Girard, L., Lockwood, W. W., Lam, W. L. & Minna, J. D. Lung cancer cell lines as tools for biomedical discovery and research. J. Natl Cancer Inst. 102, 1310–1321 (2010).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Li, L. et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat. Commun. 5, 5469 (2014).
Marino, S., Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Toker, A. & Cantley, L. C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387, 673–676 (1997).
Hicks, S. N. et al. General and versatile autoinhibition of PLC isozymes. Mol. Cell 31, 383–394 (2008).
Bunney, T. D. et al. Structural and functional integration of the PLCγ interaction domains critical for regulatory mechanisms and signaling deregulation. Structure 20, 2062–2075 (2012).
Rutter, G. A. et al. Subcellular imaging of intramitochondrial Ca2+ with recombinant targeted aequorin: significance for the regulation of pyruvate dehydrogenase activity. Proc. Natl Acad. Sci. USA 93, 5489–5494 (1996).
Jouaville, L. S., Pinton, P., Bastianutto, C., Rutter, G. A. & Rizzuto, R. Regulation of mitochondrial ATP synthesis by calcium: evidence for a long-term metabolic priming. Proc. Natl Acad. Sci. USA 96, 13807–13812 (1999).
Rimessi, A. et al. Interorganellar calcium signaling in the regulation of cell metabolism: a cancer perspective. Semin. Cell. Dev. Biol. 98, 167–180 (2020).
Rana, R. S. & Hokin, L. E. Role of phosphoinositides in transmembrane signaling. Physiol. Rev. 70, 115–164 (1990).
Rhee, S. G. & Bae, Y. S. Regulation of phosphoinositide-specific phospholipase C isozymes. J. Biol. Chem. 272, 15045–15048 (1997).
Rimessi, A., Giorgi, C., Pinton, P. & Rizzuto, R. The versatility of mitochondrial calcium signals: from stimulation of cell metabolism to induction of cell death. Biochim. Biophys. Acta 1777, 808–816 (2008).
Guzy, R. D. et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1, 401–408 (2005).
Dada, L. A. et al. Hypoxia-induced endocytosis of Na,K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-ζ. J. Clin. Invest. 111, 1057–1064 (2003).
Tarasov, A. I., Griffiths, E. J. & Rutter, G. A. Regulation of ATP production by mitochondrial Ca2+. Cell Calcium 52, 28–35 (2012).
Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011).
Giorgi, C., Marchi, S. & Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 19, 713–730 (2018).
Negre-Salvayre, A., Coatrieux, C., Ingueneau, C. & Salvayre, R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br. J. Pharmacol. 153, 6–20 (2008).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Wilson, W. R. & Hay, M. P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 11, 393–410 (2011).
Dewhirst, M. W., Cao, Y. & Moeller, B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat. Rev. Cancer 8, 425–437 (2008).
Wang, X. T., McCullough, K. D., Wang, X. J., Carpenter, G. & Holbrook, N. J. Oxidative stress-induced phospholipase C-γ1 activation enhances cell survival. J. Biol. Chem. 276, 28364–28371 (2001).
Sala, G. et al. Phospholipase Cγ1 is required for metastasis development and progression. Cancer Res. 68, 10187–10196 (2008).
Thomas, S. M. et al. Epidermal growth factor receptor-stimulated activation of phospholipase Cγ-1 promotes invasion of head and neck squamous cell carcinoma. Cancer Res. 63, 5629–5635 (2003).
Turner, T., Epps-Fung, M. V., Kassis, J. & Wells, A. Molecular inhibition of phospholipase cγ signaling abrogates DU-145 prostate tumor cell invasion. Clin. Cancer Res. 3, 2275–2282 (1997).
Tang, W. et al. Oncogenic role of phospholipase C-γ1 in progression of hepatocellular carcinoma. Hepatol. Res. 49, 559–569 (2019).
Ackerman, D. et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 24, 2596–2605.e5 (2018).
Zhang, X. et al. Inhibition of intracellular lipolysis promotes human cancer cell adaptation to hypoxia. eLife 6, e31132 (2017).
Liu, L. et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015).
Teppo, H. R., Soini, Y. & Karihtala, P. Reactive oxygen species-mediated mechanisms of action of targeted cancer therapy. Oxid. Med. Cell. Longev. 2017, 1485283 (2017).
Gresset, A., Hicks, S. N., Harden, T. K. & Sondek, J. Mechanism of phosphorylation-induced activation of phospholipase C-γ isozymes. J. Biol. Chem. 285, 35836–35847 (2010).
De Stefani, D., Raffaello, A., Teardo, E., Szabo, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009).
Stewart, S. A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003).
Wang, L., Jin, Q., Lee, J. E., Su, I. H. & Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl Acad. Sci. USA 107, 7317–7322 (2010).
Phelps, R. M. et al. NCI-Navy medical oncology branch cell line data base. J. Cell Biochem. Suppl. 24, 32–91 (1996).
Coughlan, A. M. et al. Myeloid engraftment in humanized mice: impact of granulocyte-colony stimulating factor treatment and transgenic mouse strain. Stem Cells Dev. 25, 530–541 (2016).
Shultz, L. D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005).
Ramadori, G. et al. Diet-induced unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell Metab. 21, 117–125 (2015).
Majka, S. M. et al. Analysis and isolation of adipocytes by flow cytometry. Methods Enzymol. 537, 281–296 (2014).
Bonora, M. et al. Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat. Protoc. 8, 2105–2118 (2013).
Padanad, M. S. et al. Fatty acid oxidation mediated by Acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 16, 1614–1628 (2016).
Traynor-Kaplan, A. et al. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim. Biophys. Acta 1862, 513–522 (2017).
Acknowledgements
We wish to thank J. Sondek for providing the rat PLCγ1 plasmid (UNC Center for Structural Biology, Chapel Hill, USA). The NSCLC cell lines were provided by J. D. Minna (UTSW medical center, USA). We thank A. Traynor-Kaplan (University of Washington, USA) for help with mass spectrometry analysis. We thank G. Ramadori (University of Geneva, Switzerland) for critically reading the manuscript. This study was supported by the German Research Council (DFG; HE6233/4-1 to F.H.H. and SCHN15561-1 to T.M.S.), the Thuringian state program ProExzellenz (RegenerAging-FSU-I-03/14) of the Thuringian Ministry for Research (TMWWDG; to F.H.H.), local funds from the University of Ferrara (FIR-2017), the Italian Ministry of Health (GR-2016-02364602) and the Italian Ministry of Education, University and Research (PRIN Grant 2017XA5J5N) (to A.R.), the Italian Association for Cancer Research (AIRC, IG-18624), Telethon (GGP11139B), the Italian Ministry of Education, University and Research (PRIN Grant 2017E5L5P3), local funds from the University of Ferrara (to P.P.), and the Swiss National Science Foundation (#PP00P3_163929) Professorship (to G.K.).
Author information
Authors and Affiliations
Contributions
G.K. and M.S. conceived and designed the experiments. M.S., M.R.S., C.P. and G.K. performed and analysed the data. F.H.H. and T.M.S. generated and provided the Plcg1fl/fl mice. S.S.P., L.B. and S.A.B. (pathologists) and R.S. (surgeon) provided the human lung tissue samples, and S.A.B. performed the PLCγ1 staining grading. A.R. and P.P. performed the Ca2+ measurements. J.B. and S.F. performed the Seahorse experiments. G.K. supervised the study. G.K. and M.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Hypoxia suppresses PLCγ1 and alters the fatty acid composition of phosphoinositides.
a-d, Total PI (a), PI4P (b), PI(4,5)P2 (c) and PI(3,4,5)P3 (d) peak areas of A549 cells in normoxia and upon shift to hypoxia (1% O2) for 48h measured by ultra-performance liquid chromatography - tandem mass spectrometry; n = 4/group. e, Schematic representation of phosphoinositol signalling and the metabolic fate of PI(4,5)P2. PM: plasma membrane. PI: phosphatidylinositol, PI4P: phosphatidylinositol-4-phosphate, PtdIns(4,5)P2: phosphatidylinositol-4,5-biphosphate, PI(3,4,5)P3: phosphatidylinositol-3,4,5-trisphosphate. f, HIF1a mRNA expression level in A549 cells (relative to normoxia control at 24h). Cells were transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against HIF1α and incubated in normoxia or hypoxia for the indicated time; n = 3. g, Heatmap display of PI(4,5)P2 lipid alterations (fold change). A549 cells were transduced with either an empty vector control (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1, treated with doxycycline for 48h and treated as in (a); n = 4/group. h, Peak areas of the indicated PI(4,5)P2 lipid species in A549 cells from (g); n = 4/group. i, Total PI(4,5)P2 peak areas in A549 cells treated as in (g); n = 4/group.All graphical data are presented as mean ± SD. Statistical analyses were done using two-tailed unpaired Student’s t test or one-way ANOVA; n, number of biologically independent samples. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Statistical source data are provided in Source Data Extended Data Fig. 1.
Extended Data Fig. 2 PLCγ1 suppression promotes cancer cell proliferation during hypoxia.
a, Immunoblot analysis of the indicated targets in H358 and A427 cells transduced with either an empty vector control (Tet-pLKO-puro, shControl) or 2 doxycycline-inducible shRNAs against PLCγ1. Cells were incubated in the presence of doxycycline for 72h in normoxia before performing the immunoblot analysis. b,c, Relative cell number of A549, H358 and A427 cells transduced with either an empty vector control (Tet-pLKO-puro, shControl), or a doxycycline-inducible shRNA against PLCγ1 (as indicated). After this, cells were plated and incubated for the indicated time in normoxia or hypoxia; n = 3. d, Immunoblot analysis of the indicated targets in A549 cells transduced as in (b). Cells were then incubated in the presence of doxycycline and transfected with either pcDNA3.1 empty vector or pcDNA3.1-rPLCγ1 (shRNA-resistant PLCγ1) and incubated for 72h in normoxia or hypoxia. e, Immunoblot analysis of the indicated targets in A549 cells transduced as in (a). f,g, Immunoblot analysis of the indicated targets (f) and cell proliferation assay (g) of A549 cells treated as indicated in normoxia or hypoxia; n = 3.All graphical data are presented as mean ± SD. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 PLCγ1 suppression promotes cancer cell survival during hypoxia.
a, Representative panels of Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis (left) and relative quantification of Annexin V (AV) + Annexin V/PI (AV/PI)-positive cells (right) in A549 cells transduced with either an empty vector control (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1, incubated without/with doxycycline for 48h and moved in normoxia or hypoxia for 72h; n = 3. b, Caspase-3 activity assay in A549 cells transduced as in (a). Cells were then transfected with either pcDNA3.1 empty vector or pcDNA3.1-rPLCγ1 (shRNA-resistant PLCγ1), and incubated in normoxia or hypoxia for 48h. Staurosporine (STA, 100nM) is positive control of caspase-dependent death; n = 3. c, Quantification of Annexin V (AV) + Annexin V/PI (AV/PI)-positive cells by flow cytometry in A549 cells transduced as in (a). Cells were then treated with Q-VD-OPH (QVD), a pan-caspase inhibitor for 24h; n = 3. d, Representative panels of Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis (left) and relative quantification of Annexin V (AV) + Annexin V/PI (AV/PI)-positive cells (right) in A549 cells transduced with either pcDNA3.1 empty vector or pcDNA3.1-PLCγ1 and incubated in hypoxia for 72h; n = 3. e, Immunoblot analysis of the indicated targets in MEF cells with the indicated genotype in normoxia or hypoxia. The samples derive from the same experiment and the gel/blot were processed in parallel. f, Representative panels of Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis of MEF cells with the indicated genotype. Graphical data are mean ± SD. Statistical analyses were done using two-tailed unpaired Student’s t test or one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 PLCγ1 suppression decreases mitochondrial respiration and enhances cancer cell glycolytic capacity in human NSCLC cells.
a, Immunoblot analysis of the indicated targets in A549 cells transduced with either an empty vector control (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1, incubated without/with doxycycline for 48h and further incubated for 48h in normoxia or hypoxia (3% O2). b-e, Graphs showing oxygen consumption rate (OCR, b and c) and extracellular acidification rate (ECAR, d and e) of A549 cells transduced and treated as in (a); n replicates/group: (b) = 8, (c) = 7 normoxia/8 hypoxia, (d) = 9 normoxia/ 8 hypoxia, (e) = 8. Panels (c and e) are treated without/with doxycycline and serve as a control for the doxycycline effect on cells. f, Bar graph showing ECAR parameters in A549 cells transduced as in (a). Cells were transfected with either pcDNA3.1 empty vector or pcDNA3.1-rPLCγ1 (shRNA-resistant PLCγ1) to rescue the shPLCγ1 #2 and incubated for 48h in normoxia or hypoxia; n = 8. g-h, Oxygen consumption rate (g) and extracellular acidification rate (h) parameters of A549 cells transfected with either pcDNA3.1 empty vector or pcDNA3.1-PLCγ1 and incubated for 48h in normoxia or hypoxia; (g) n = 8 for normoxia and 6 for hypoxia groups, (h) n = 7.OCR was determined during sequential treatments with oligomycin, FCCP and rotenone/antimycin (AA). ECAR was determined during sequential treatments with glucose (Glc), oligomycin and 2 deoxyglucose (2DG).Graphical data are mean ± SD. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 PLCγ1 suppression decreases mitochondrial respiration and enhances cancer cell glycolytic capacity in MEFs.
a, Immunoblot analysis of the indicated targets in MEF cells with the indicated genotype transfected with either pcDNA3.1-HA-LIC empty vector or pcDNA3.1-HA-LIC-PLCγ1. 24h later, cells were moved in hypoxia (3% O2) and incubated for additional 48h. b, Oxygen consumption rate (OCR) graph of MEF cells of the indicated genotypes. Cells were incubated for 48h in normoxia or hypoxia (3% O2) before seahorse experiment. OCR was determined during sequential treatments (indicated with arrows) with oligomycin, FCCP and rotenone/antimycin (AA) at the indicated time; n = 3 for the normoxia groups and n = 6 for the hypoxia groups. c, Graph showing extracellular acidification rate (ECAR) of MEF cells with the indicated genotype transduced as in (a). ECAR was determined during sequential treatments (indicated with arrows) with glucose (Glc), oligomycin and 2 deoxyglucose (2DG) at the indicated time; n = 6. d, Flow cytometry panels showing mean fluorescent intensity of A549 cells from experiment reported in main Fig. 3i. A549 cells were previously transduced with either an empty vector control (Tet-pLKO-puro, shControl) or 2 different doxycycline-inducible shRNAs against PLCγ1 (shPLCγ1 #1, shPLCγ1 #2), incubated in the presence of doxycycline for 48h, incubated for additional 48h in normoxia or hypoxia (1% O2), stained with LipidTOX and analyzed by flow cytometry. Sh#1: shPLCγ1 #1, Sh#2: shPLCγ1 #2; n = 3. Graphical data are mean ± SD; n, number of biologically independent samples. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 PLCγ1 suppression depletes mitochondrial ROS through impairment of Ca2+ entry into the mitochondria.
a, Flow cytometry histograms (left) and quantification of DHR (green, right) of A549 cells transduced with an empty vector (Tet-pLKO-puro, shControl) and incubated without/with doxycycline (DOX) for 48h, incubated for additional 48h in normoxia or hypoxia and stained with DHR for flow-cytometry. H2O2: positive control. DHR: Dihydrorhodamine. Normo: normoxia. Hypo: hypoxia. n = 3. b, MCU mRNA levels in A549 cells transduced with an empty vector (Tet-pLKO-puro, shControl) or 2 doxycycline-inducible shRNAs against MCU and incubated with doxycycline for 72h; n = 3. c, Immunoblot analysis of the indicated targets in A549 cells transduced with an empty vector (Tet-pLKO-puro, shControl) or 2 doxycycline-inducible shRNA against PLCγ1, transfected with either pcDNA3.1 empty vector or pcDNA3.1-MCU-flag plasmid and incubated in normoxia or hypoxia for 48h. d, Representative flow cytometry histograms of DHR (green) mean fluorescent intensity of A549 cells from experiment reported in main Fig. 4f. Sh#2: shPLCγ1 #2. e, Representative flow cytometry histograms (left) and quantification of DHR (green) of A549 cells transduced with an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1. Cells were then treated with doxycycline for 24h, transfected with either pcDNA3.1 empty vector or pcDNA3.1-MCU-flag plasmid and moved to hypoxia for additional 48h before staining cells with DHR for analysis. DHR: Dihydrorhodamine. Sh#1: shPLCγ1 #1; n = 3. f, Cytosolic (left) and mitochondrial (right) Ca2+ response of A549 cells transduced as in (c), transfected with appropriate targeted-aequorin and histamine-induced Ca2+ was measured 48h after incubation in normoxia or hypoxia; n/group = cytosolic normoxia 7, 8, 7, 7, 8, 7/cytosolic hypoxia 8, 6, 7, 8, 7, 8. n/group = mitochondrial normoxia 11, 7, 9, 10, 8, 6/mitochondrial hypoxia 7, 10, 9, 9, 9, 11.Graphical data are mean ± SD. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 6.
Extended Data Fig. 7 PLCγ1 suppression enhances cell proliferation and decreases cell death in hypoxia through impairment of Ca2+ entry into the mitochondria.
a, Immunoblot analysis of the indicated targets in MEFs generated from a LSL-KrasG12D/WT;p53flox/flox;Plcg1wt/wt and LSL-KrasG12D/WT;p53flox/flox;Plcg1flox/flox mouse model, stably transduced with Cre recombinase (pMSCV-hygro-Cre) and transfected with either pcDNA3.1 empty vector or pcDNA3.1- MCU-flag plasmid and moved to hypoxia (1% O2) for additional 48h. b,c, Oxygen consumption rate (b) and extracellular acidification rate (c) of A549 cells transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1, treated with doxycycline for 24h, transfected with either pcDNA3.1 empty vector or pcDNA3.1-MCU-flag plasmid and incubated for 48h in hypoxia (3% O2). OCR was determined during sequential treatments (indicated with arrows) with oligomycin, FCCP and rotenone/antimycin (AA) at the indicated time. ECAR was determined during sequential treatments (indicated with arrows) with glucose (Glc), oligomycin and 2 deoxyglucose (2DG) at the indicated time; n = 5. d, Fatty acid β-oxidation of A549 cells transduced and treated as in (b); n = 4. e, Relative cell number of A549 cells transduced and treated as in (b); n = 3. Normoxia: * p = 0.03, hypoxia: * p = 0.014, *** p = 0.0005. f, Representative Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis panels of A549 cells from experiment reported in main Fig. 5e; n = 3. g, Representative Annexin V-Atto 633/Propidium iodide (PI) flow cytometry analysis panels of MEF cells from experiment reported in main Fig. 5f; n = 3. Graphical data are mean ± SD. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data and unprocessed immunoblots are provided in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 PLCγ1 suppression reduces hypoxia-induced lipid peroxidation.
a, Confocal microscopy of A549 cells transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1 (shPLCγ1 #1) and incubated in the presence of doxycycline for 48h. Cells were then incubated for 48h in normoxia or hypoxia (1% O2), stained with BODIPY 581/591 C11 (green: oxidized lipids / red: non-oxidized lipids), LipidTOX (grey) to mark lipid droplets and Hoechst (nuclei). White squares represent magnified areas on the right. Scale bars: 20 μm. b, Quantification of lipid peroxidation expressed as percent of oxidized lipids from (a); n/group = 13, 13, 12, 14. c, Quantification of oxidized lipids, expressed as percent, localized in lipid droplets from (a); n/group = 13, 13, 13, 14. d, Relative cell number of A549 (left) and H358 (right) cells transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1 (shPLCγ1 #1), incubated with doxycycline for 48h and treated as indicated with the DGAT1 inhibitor (T863). The graphs represent day 6 of the cell proliferation assay; n = 3. e, Lipid droplet quantification (relative to control) of A549 and H358 cells treated as in (d), stained with LipidTOX (far red) and analyzed by flow cytometry; n = 3.Graphical data are mean ± SD except the panels b and c that are mean ± SEM. Statistical analyses were done using one-way ANOVA; n, number of biologically independent samples. **** p < 0.0001. Statistical source data are provided in Source Data Extended Data Fig. 8.
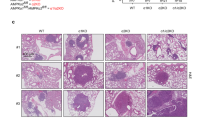
Extended Data Fig. 9 PLCγ1 deletion accelerates lung tumorigenesis in mice.
a, Representative hematoxylin & eosin (H&E) staining of LSL-KrasG12D/WT;p53flox/flox;Plcg1wt/wt, LSL-KrasG12D/WT;p53flox/flox;Plcg1wt/flox and LSL-KrasG12D/WT;p53flox/flox;Plcg1flox/flox mouse lung sections, 10 weeks after Cre induction. Scale bar: 5000μm. b, Representative immunohistochemistry images showing Ki67-positive cell staining in mouse lung tumors from LSL-KrasG12D/WT;p53flox/flox;Plcg1wt/wt, KrasG12D/WT;p53flox/flox;Plcg1wt/flox and LSL-KrasG12D/WT;p53flox/flox;Plcg1flox/flox mice 10 weeks after Cre induction. This is related to main Fig. 7e.
Extended Data Fig. 10 Low PLCγ1 levels in human lung adenocarcinomas correlate with poor patient survival.
a, Antibody validation for IHC against PLCγ1 on human tumor tissue microarray. A549 cells previously transduced with either an empty vector (Tet-pLKO-puro, shControl) or a doxycycline-inducible shRNA against PLCγ1 (shPLCγ1 #1) and incubated in the presence of doxycycline for 48h before paraffin embedding. Subsequently IHC was performed as for TMA staining (described in materials and methods). The arrow heads indicate PLCγ1-positive cells shown at higher magnification in the inset b, Representative images of the human lung adenocarcinoma TMA showing the staining intensity with the matched scoring by the pathologist upon immunohistochemistry against PLCγ1. Score 0-I was considered low staining intensity, II was considered moderate and score III was considered high staining intensity.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables 1 and 2
Supplementary Table 1: detailed reagents and tools information. Supplementary Table 2: list of the patient age, gender and corresponding PLC 0-3 score (used in the TMA).
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed immunoblots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed immunoblots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed immunoblots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed immunoblots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed immunoblots
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed immunoblots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed immunoblots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed immunoblots.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Saliakoura, M., Rossi Sebastiano, M., Pozzato, C. et al. PLCγ1 suppression promotes the adaptation of KRAS-mutant lung adenocarcinomas to hypoxia. Nat Cell Biol 22, 1382–1395 (2020). https://doi.org/10.1038/s41556-020-00592-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-00592-8
This article is cited by
-
Self-assembly of DNA nanostructure containing cell-specific aptamer as a precise drug delivery system for cancer therapy in non-small cell lung cancer
Journal of Nanobiotechnology (2022)
-
Restriction of extracellular lipids renders pancreatic cancer dependent on autophagy
Journal of Experimental & Clinical Cancer Research (2022)