Abstract
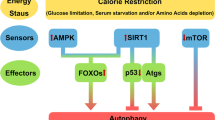
SIRT1 (Sir2) is an NAD+-dependent deacetylase that plays critical roles in a broad range of biological events, including metabolism, the immune response and ageing1,2,3,4,5. Although there is strong interest in stimulating SIRT1 catalytic activity, the homeostasis of SIRT1 at the protein level is poorly understood. Here we report that macroautophagy (hereafter referred to as autophagy), a catabolic membrane trafficking pathway that degrades cellular components through autophagosomes and lysosomes, mediates the downregulation of mammalian SIRT1 protein during senescence and in vivo ageing. In senescence, nuclear SIRT1 is recognized as an autophagy substrate and is subjected to cytoplasmic autophagosome–lysosome degradation, via the autophagy protein LC3. Importantly, the autophagy–lysosome pathway contributes to the loss of SIRT1 during ageing of several tissues related to the immune and haematopoietic system in mice, including the spleen, thymus, and haematopoietic stem and progenitor cells, as well as in CD8+CD28− T cells from aged human donors. Our study reveals a mechanism in the regulation of the protein homeostasis of SIRT1 and suggests a potential strategy to stabilize SIRT1 to promote productive ageing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA sequencing data were referenced to the Gene Expression Omnibus under accession numbers GSE52848 and GSE53356. The mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD020081. The authors declare that the data that support the findings of this study are available within the manuscript. No restriction on data availability applies. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Hall, J. A., Dominy, J. E., Lee, Y. & Puigserver, P. The sirtuin family’s role in aging and age-associated pathologies. J. Clin. Invest. 123, 973–979 (2013).
Haigis, M. C. & Sinclair, D. A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 (2010).
Lin, S. J., Defossez, P. A. & Guarente, L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 (2000).
Tissenbaum, H. A. & Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 (2001).
Rogina, B. & Helfand, S. L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl Acad. Sci. USA 101, 15998–16003 (2004).
Dang, W. et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 (2009).
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Chimenti, C. et al. Senescence and death of primitive cells and myocytes lead to premature cardiac aging and heart failure. Circ. Res. 93, 604–613 (2003).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Huang, J. et al. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS ONE 3, e1710 (2008).
Sasaki, T., Maier, B., Bartke, A. & Scrable, H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell 5, 413–422 (2006).
Rai, T. S. et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 28, 2712–2725 (2014).
Amaravadi, R. K. & Winkler, J. D. Lys05: a new lysosomal autophagy inhibitor. Autophagy 8, 1383–1384 (2012).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Narita, M. et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011).
Gerland, L. M. et al. Association of increased autophagic inclusions labeled for β-galactosidase with fibroblastic aging. Exp. Gerontol. 38, 887–895 (2003).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Kerppola, T. K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008).
Birgisdottir, A. B., Lamark, T. & Johansen, T. The LIR motif—crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 (2013).
Huang, R. et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 (2015).
Noda, N. N. et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 (2008).
Kraft, L. J., Nguyen, T. A., Vogel, S. S. & Kenworthy, A. K. Size, stoichiometry, and organization of soluble LC3-associated complexes. Autophagy 10, 861–877 (2014).
Alemu, E. A. et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287, 39275–39290 (2012).
Skytte Rasmussen, M. et al. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy 13, 834–853 (2017).
Johansen, T. et al. Methods for studying interactions between Atg8/LC3/GABARAP and LIR-containing proteins. Methods Enzymol. 587, 143–169 (2017).
Dai, H. et al. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat. Commun. 6, 7645 (2015).
Davenport, A. M., Huber, F. M. & Hoelz, A. Structural and functional analysis of human SIRT1. J. Mol. Biol. 426, 526–541 (2014).
Escande, C. et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Invest. 120, 545–558 (2010).
Rimmele, P. et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Rep. 3, 44–59 (2014).
Huff, W. X., Kwon, J. H., Henriquez, M., Fetcko, K. & Dey, M. The evolving role of CD8+CD28− immunosenescent T cells in cancer immunology. Int. J. Mol. Sci. 20, 2810 (2019).
Jeng, M. Y. et al. Metabolic reprogramming of human CD8+ memory T cells through loss of SIRT1. J. Exp. Med. 215, 51–62 (2018).
Zu, Y. et al. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ. Res. 106, 1384–1393 (2010).
Chen, H. et al. SIRT1 ameliorates age-related senescence of mesenchymal stem cells via modulating telomere shelterin. Front. Aging Neurosci. 6, 103 (2014).
Fernandez, A. F. et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018).
Hayakawa, T. et al. SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS ONE 10, e0116480 (2015).
Montecino-Rodriguez, E., Berent-Maoz, B. & Dorshkind, K. Causes, consequences, and reversal of immune system aging. J. Clin. Invest. 123, 958–965 (2013).
Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013).
Dou, Z. et al. Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol. Cell 50, 29–42 (2013).
Sun, L. & Fang, J. Macromolecular crowding effect is critical for maintaining SIRT1’s nuclear localization in cancer cells. Cell Cycle 15, 2647–2655 (2016).
Acknowledgements
We thank J. Zhao for help with confocal microscopy and L. Zhang for help with cell sorting by flow cytometry. We thank P. Klein, E. J. Wherry, F. Mourkioti and M. G. Vizioli for their help with the mouse experiments; R. Marmorstein for help with the SIRT1–LC3 interaction mapping experiments; and N. Raman and the Gladstone Flow Cytometry Core for assistance with FACS. C.X. acknowledges the Glenn/AFAR Scholarship for Research in the Biology of Aging. Z.D. is supported by NIH grant no. K99AG053406. S.L.B. and P.D.A. are supported by NIH grant no. P01AG031862. S.L.B. acknowledges the Glenn Foundation Award for Research in Biological Mechanisms of Aging.
Author information
Authors and Affiliations
Contributions
C.X., Z.D. and S.L.B. conceived the project. C.X. performed most of the experiments. Z.D. and L.W. performed cell-culture experiments. L.W., P.F. and M.O. performed human T-cell experiments. V.C., J.J. and W.T. performed HSPC isolation. M.N., J.D.W. and R.A. contributed autophagy reagents. T.J. and G.E. performed peptide arrays. C.L. and B.A.G. performed mass spectrometry analysis. C.B. and P.D.A. contributed to the mouse experiments. C.X., L.W., P.D.A., Z.D. and S.L.B. contributed to the experimental design. C.X., L.W., Z.D. and S.L.B. wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of SIRT1 role and mRNA levels in cellular senescence.
a, Western blot showing SIRT1 expression in IMR90 cells stably expressing LPC vector or LPC-HA-SIRT1 construct; n = 3 independent experiments. b, β-gal-stained cells at day 9 post etoposide treatment were imaged by microscopy; n = 2 independent experiments. OE: overexpression. c, Percentages of β-gal staining-positive cells were quantified at indicated days after etoposide treatment. Data are mean ± s.d.; more than 500 cells and 5 fields were counted; two-way ANOVA with Sidak’s multiple comparisons test (p values). d, Western blot showing SIRT1 expression in primary BJ fibroblasts treated with 100 μM etoposide for 48 h in time course as indicated; n = 2 independent experiments. e, Western blot showing SIRT1 expression in IMR90 cells cultured at 100% confluency for indicated days; n = 2 independent experiments. f-i, RT-qPCR time course analysis of proliferating and senescent IMR90 cells induced by overexpressing ER:HRasV12 treated with 4OHT (f), by etoposide for 48 h (g) and BJ cells treated with etoposide for 48 h (h), and in IMR90 replicative senescent cells (i). Data were normalized to GAPDH; the bar indicates the average of three technical replicates. PD, population doubling. j, SIRT1 deacetylation activity assay of endogenous SIRT1 protein immunoprecipitated from extracts of proliferating or senescent IMR90 cells expressing inducible hairpins of shNTC and shAtg7. For senescent cells, cells were induced by Dox for 5 days, and then were subjected to etoposide treatment for 48h; Cells at Day 8 after etoposide treatment were harvested for analysis. For proliferating cells, cells were induced by Dox for 5 days and then were harvested for analysis. Proteins loaded were analysed by western blotting. This experiment has been repeated for two times. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 2 Characterization of the SIRT1–LC3 interaction.
a, IP of extracts from proliferating and senescent IMR90 cells. Quantification: SIRT1 IP bands were normalized to LC3 IP and SIRT1 input bands. Mean ± s.d.; n = 6 independent experiments; paired two-tailed Students’ t-test. b, IP of extracts from proliferating and contact-inhibited IMR90 cells (at 100% confluency for 8 days). This experiment has been repeated once. Excessive beads and antibodies were used in the IP to capture nearly 100% of LC3 protein in the lysates. Flow: flow-through. c, IP of extracts from proliferating and contact-inhibited cells; n = 3 independent experiments. d, IP of nuclear extracts from proliferating and senescent cells. Excessive beads and antibodies were used to capture nearly 100% of LC3 protein in the lysates. This experiment has been repeated once. Flow: flow-through. e, IP of nuclear extracts from proliferating and senescent cells; n = 2 independent experiments. In a,d,e, senescent cells were harvested at day 8 after etoposide treatment. f, Western blotting of nuclear (Nuclear) and cytoplasmic (Cyto) extracts from proliferating and senescent cells; n = 2 independent experiments. g, Endogenous LC3 IP of IMR90 cell extracts with or without protein phosphatase Lambda treatment. Quantification: SIRT1 IP bands were normalized to LC3 IP and SIRT1 input bands. Mean ± s.d.; paired one-tailed Students’ t-test; n = 5 independent experiments. h-i, Mass spectrometry analysis of SIRT1 immunoprecipitated from proliferating and senescent IMR90 cells. h, Boxplot showing the peptide intensity distribution of SIRT1. N = 54 peptides; p value = 0.25; unpaired two-tailed Student’s t-test. The median of the data was indicated as the line in the box, and edges stand for the 25th/75th percentile. i, Phosphorylated peptides identified by mass spec and their phosphorylation levels in proliferating and senescence states. AA: amino acid. Source data are provided.
Extended Data Fig. 3 Characterization of the SIRT1 deacetylation role in starvation and senescence.
a-c, IMR90 cells that undergo CRISPR/Cas9-mediated gene inactivation of non-targeting control (PRM1, as PRM1 is involved in spermatogenesis and is not expressed in IMR90 cells) or SIRT1 were analysed under starvation and senescence conditions. This experiment has been repeated for two times. a, Cells were analysed by western blotting. b, Cells were subjected to 250 μM Torin 1 and 5 μM Lys05 treatment for 24 h, and analysed by western blotting. Relative LC3-II intensities to GAPDH were quantified. c, Cells at day 6 after etoposide-treated senescence were subjected to 2 μM or 5 μM Lys05 treatments for 24 h, and analysed by western blotting. Relative LC3-II intensities to GAPDH were quantified. d-e, IMR90 cells under proliferating, starvation (Torin 1 250 μM for 24 h) and senescence (induced by etoposide treatment, harvested at day 7) conditions were stained with LC3 antibody and analysed. d, Cells were imaged by confocal microscopy. Scale bar: 10 μm. e, Percentages of cells with nuclear LC3 signals were quantified. Starv: starvation. Mean ± s.d.; more than 500 cells were counted; each data point (n) represents cells in 10 random fields, n = 5 for all conditions; one-way ANOVA coupled with Turkey’s multiple comparisons test. f, IMR90 were treated as indicated ways for 24 h and analysed by western blotting; n = 2 independent experiments. CT: control. A.A.: amino acids. 2-DG: treatment of 10 mM 2-DG. Torin 1: treatment of 250 μM Torin 1. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 4 Characterization of SIRT1 mutants and peptides.
a, Information of potential SIRT1–LC3 interaction regions identified in the peptide array as in Fig. 4a, and the corresponding synthetic peptides and mutants. Key amino acid residues are labelled in red. Potential region: LC3-binding regions on SIRT1 identified in the peptide array as in Fig. 4a. Peptide region: synthetic peptides tested in the peptide competition IP as in Fig. 4b. Peptide competition: results of the peptide competition IP as in Fig. 4b. Substitution generated: SIRT1 mutants tested in the IP as in Fig. 4c. b, SIRT1 deacetylation activity assay of SIRT1 WT or WV mutant immunoprecipitated from extracts of HEK293T expressing corresponding HA-tagged constructs. Proteins loaded were analysed by western blotting. This experiment has been repeated for two times. c, IP of HEK293T cells expressing HA-SIRT1 and Flag-LC3 constructs; n = 2 independent experiments. Cells were pre-treated with 20 μM resveratrol for 6 h. d, IMR90 cells at day 6 after etoposide-initiated senescence were subjected to treatment with 20 μM resveratrol for 48 h; n = 2 independent experiments. Cells were then harvested for western blotting. e, HEK293T cells expressing HA-SIRT1 and Flag-LC3 were treated with 20 μM resveratrol for 6 h, and were then harvested for SIRT1 activity assay. This experiment has been repeated for two times. f, SIRT1 deacetylation activity assay of SIRT1 WT or I347A mutant immunoprecipitated from extracts of HEK293T expressing corresponding HA-tagged constructs. Proteins loaded were analysed by western blotting; n = 2 independent experiments. g, IP of HEK293T cell lysates expressing Flag-tagged LC3 and HA-tagged SIRT1 WT or I347A, WV+I347A, or WV mutants. This experiment has been repeated for two times. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 5 Analysis of SIRT1 in mouse tissues and HSPCs.
a,b, Thymus from young (2-4 months) and aged (19-26 months) C57BL/6 mice were lysed and analysed by western blotting (a) and RT-qPCR (b); n = 3 biologically independent animals in each group. RT-qPCR data were normalized to 18S; mean ± s.e.m.; unpaired two-tailed Students’ t-test. c. Indicated organs and tissues of young (3 months) and aged (19 months) mice were dissected and analysed by western blotting; n = 2 independent experiments. SE: short exposure; LE: long exposure. d,e, Young (3 months) mice were fed or fasted for 24 h. Spleens (d) and Testes (e) were harvested for western blotting; n = 2 biologically independent animals in each group. f,g, Young (2-4 months) mice were subjected to daily i.p. injection of 10 mg/kg Lys05 in PBS or PBS control in 100 μL volume for two weeks. Spleens (f) and testes (g) were analysed by western blotting. Western blot quantification: SIRT1 bands were normalized to GAPDH bands. For spleens, data are mean ± s.e.m.; control group n = 3 animals, Lys05 group n = 5 animals; two-tailed Mann-Whitney test. For testes, data are mean ± s.e.m.; n = 4 animals; two-tailed Mann-Whitney test. h, Representative flow cytometry plots of cell sorting of lineage-depleted bone marrow cells from young and aged mice to isolate Lin-Sca-1+c-Kit+ cells (HSPC populations). Boxes indicate cell populations isolated. i, HSPC populations were isolated from young (2-4 months) mice, cultured with or without 2 μM Lys05 for 24 hours and analysed by western blotting. This experiment has been repeated once. Statistical information and unprocessed blots are provided as source data.
Supplementary information
Supplementary Information
Supplementary Fig. 1. Gating strategies used for cell sorting.
Supplementary Table 1
Antibody list.
Source data
Source Data Fig. 1
Statistical source data
Unprocessed Blots Fig. 1
Unprocessed western blots
Source Data Fig. 2
Statistical source data
Unprocessed Blots Fig. 2
Unprocessed western blots
Source Data Fig. 3
Statistical source data
Unprocessed Blots Fig. 3
Unprocessed western blots
Source Data Fig. 4
Statistical source data
Unprocessed Blots Fig. 4
Unprocessed western blots
Source Data Fig. 5
Statistical source data
Unprocessed Blots Fig. 5
Unprocessed western blots
Source Data Extended Data Fig. 1
Statistical source data
Unprocessed Blots Extended Data Fig. 1
Unprocessed western blots
Source Data Extended Data Fig. 2
Statistical Source Data
Unprocessed Blots Extended Data Fig. 2
Unprocessed western blots
Source Data Extended Data Fig. 3
Statistical source data
Unprocessed Blots Extended Data Fig. 3
Unprocessed western blots
Source Data Extended Data Fig. 4
Statistical source data
Unprocessed Blots Extended Data Fig. 4
Unprocessed western blots
Source Data Extended Data Fig. 5
Statistical source data
Unprocessed Blots Extended Data Fig. 5
Unprocessed western blots
Rights and permissions
About this article
Cite this article
Xu, C., Wang, L., Fozouni, P. et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat Cell Biol 22, 1170–1179 (2020). https://doi.org/10.1038/s41556-020-00579-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-00579-5
This article is cited by
-
Sirtuins in kidney health and disease
Nature Reviews Nephrology (2024)
-
Lonicerin promotes wound healing in diabetic rats by enhancing blood vessel regeneration through Sirt1-mediated autophagy
Acta Pharmacologica Sinica (2024)
-
Pro-ferroptotic signaling promotes arterial aging via vascular smooth muscle cell senescence
Nature Communications (2024)
-
Neuronal Stem Cells from Late-Onset Alzheimer Patients Show Altered Regulation of Sirtuin 1 Depending on Apolipoprotein E Indicating Disturbed Stem Cell Plasticity
Molecular Neurobiology (2024)
-
Metformin protects human lens epithelial cells from high glucose-induced senescence and autophagy inhibition by upregulating SIRT1
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)